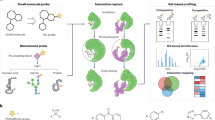
Abstract
We describe a multipurpose technology platform, termed μSCALE (microcapillary single-cell analysis and laser extraction), that enables massively parallel, quantitative biochemical and biophysical measurements on millions of protein variants expressed from yeast or bacteria. μSCALE spatially segregates single cells within a microcapillary array, enabling repeated imaging, cell growth and protein expression. We performed high-throughput analysis of cells and their protein products using a range of fluorescent assays, including binding-affinity measurements and dynamic enzymatic assays. A precise laser-based extraction method allows rapid recovery of live clones and their genetic material from microcapillaries for further study. With μSCALE, we discovered a new antibody against a clinical cancer target, evolved a fluorescent protein biosensor and engineered an enzyme to reduce its sensitivity to its inhibitor. These protein analysis and engineering applications each have unique assay requirements and different host organisms, highlighting the flexibility and technical capabilities of the μSCALE platform.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Lander, E.S. Initial impact of the sequencing of the human genome. Nature 470, 187–197 (2011).
Rahman, N. Realizing the promise of cancer predisposition genes. Nature 505, 302–308 (2014).
Rhodes, D.R. & Chinnaiyan, A.M. Integrative analysis of the cancer transcriptome. Nat. Genet. 37 (suppl.) S31–S37 (2005).
Zhang, Y. et al. Time-resolved mass spectrometry of tyrosine phosphorylation sites in the epidermal growth factor receptor signaling network reveals dynamic modules. Mol. Cell. Proteomics 4, 1240–1250 (2005).
Romero, P.A. & Arnold, F.H. Exploring protein fitness landscapes by directed evolution. Nat. Rev. Mol. Cell Biol. 10, 866–876 (2009).
Kariolis, M.S. et al. An engineered Axl 'decoy receptor' effectively silences the Gas6-Axl signaling axis. Nat. Chem. Biol. 10, 977–983 (2014).
Wu, I. & Arnold, F.H. Engineered thermostable fungal Cel6A and Cel7A cellobiohydrolases hydrolyze cellulose efficiently at elevated temperatures. Biotechnol. Bioeng. 110, 1874–1883 (2013).
Alford, S.C., Ding, Y., Simmen, T. & Campbell, R.E. Dimerization-dependent green and yellow fluorescent proteins. ACS Synth. Biol. 1, 569–575 (2012).
Chen, I., Dorr, B.M. & Liu, D.R. A general strategy for the evolution of bond-forming enzymes using yeast display. Proc. Natl. Acad. Sci. USA 108, 11399–11404 (2011).
Gai, S.A. & Wittrup, K.D. Yeast surface display for protein engineering and characterization. Curr. Opin. Struct. Biol. 17, 467–473 (2007).
Lee, C.V. et al. High-affinity human antibodies from phage-displayed synthetic Fab libraries with a single framework scaffold. J. Mol. Biol. 340, 1073–1093 (2004).
Martis, E. High-throughput screening: the hits and leads of drug discovery. J. Appl. Pharm. Sci. 01, 02–10 (2011).
Povolotskaya, I.S. & Kondrashov, F.A. Sequence space and the ongoing expansion of the protein universe. Nature 465, 922–926 (2010).
Agresti, J.J. et al. Ultrahigh-throughput screening in drop-based microfluidics for directed evolution. Proc. Natl. Acad. Sci. USA 107, 4004–4009 (2010).
Ostafe, R., Prodanovic, R., Nazor, J. & Fischer, R. Ultra-high-throughput screening method for the directed evolution of glucose oxidase. Chem. Biol. 21, 414–421 (2014).
Zinchenko, A. et al. One in a million: flow cytometric sorting of single cell-lysate assays in monodisperse picolitre double emulsion droplets for directed evolution. Anal. Chem. 86, 2526–2533 (2014).
Fischlechner, M. et al. Evolution of enzyme catalysts caged in biomimetic gel-shell beads. Nat. Chem. 6, 791–796 (2014).
Romero, P.A., Tran, T.M. & Abate, A.R. Dissecting enzyme function with microfluidic-based deep mutational scanning. Proc. Natl. Acad. Sci. USA 112, 7159–7164 (2015).
Lafferty, M. & Dycaico, M.J. GigaMatrix: an ultra high-throughput tool for accessing biodiversity. J. Lab. Autom. 9, 200–208 (2004).
Fukuda, T., Shiraga, S., Kato, M. & Yamamura, S. Construction of novel single-cell screening system using a yeast cell chip for nano-sized modified-protein-displaying libraries. NanoBiotechnology 1, 105–111 (2005).
Gorris, H.H. & Walt, D.R. Mechanistic aspects of horseradish peroxidase elucidated through single-molecule studies. J. Am. Chem. Soc. 131, 6277–6282 (2009).
Liebherr, R.B., Renner, M. & Gorris, H.H. A single molecule perspective on the functional diversity of in vitro evolved β-glucuronidase. J. Am. Chem. Soc. 136, 5949–5955 (2014).
Rissin, D.M. et al. Single-molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations. Nat. Biotechnol. 28, 595–599 (2010).
Love, J.C., Ronan, J.L., Grotenbreg, G.M., van der Veen, A.G. & Ploegh, H.L. A microengraving method for rapid selection of single cells producing antigen-specific antibodies. Nat. Biotechnol. 24, 703–707 (2006).
Fitzgerald, V. et al. Exploiting highly ordered subnanoliter volume microcapillaries as microtools for the analysis of antibody producing cells. Anal. Chem. 87, 997–103 (2015).
Kovac, J.R. & Voldman, J. Intuitive, image-based cell sorting using optofluidic cell sorting. Anal. Chem. 79, 9321–9330 (2007).
Gach, P.C., Attayek, P.J., Whittlesey, R.L., Yeh, J.J. & Allbritton, N.L. Micropallet arrays for the capture, isolation and culture of circulating tumor cells from whole blood of mice engrafted with primary human pancreatic adenocarcinoma. Biosens. Bioelectron. 54, 476–483 (2014).
Waters, J.C. Accuracy and precision in quantitative fluorescence microscopy. J. Cell Biol. 185, 1135–1148 (2009).
Lemke, G. & Rothlin, C.V. Immunobiology of the TAM receptors. Nat. Rev. Immunol. 8, 327–336 (2008).
Graham, D.K., DeRyckere, D., Davies, K.D. & Earp, H.S. The TAM family: phosphatidylserine-sensing receptor tyrosine kinases gone awry in cancer. Nat. Rev. Cancer 14, 769–785 (2014).
Sidhu, S.S. et al. Phage-displayed antibody libraries of synthetic heavy chain complementarity determining regions. J. Mol. Biol. 338, 299–310 (2004).
Ai, H.-W., Baird, M.A., Shen, Y., Davidson, M.W. & Campbell, R.E. Engineering and characterizing monomeric fluorescent proteins for live-cell imaging applications. Nat. Protoc. 9, 910–928 (2014).
Shaner, N.C. et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22, 1567–1572 (2004).
Alford, S.C., Abdelfattah, A.S., Ding, Y. & Campbell, R.E. A fluorogenic red fluorescent protein heterodimer. Chem. Biol. 19, 353–360 (2012).
Ding, Y. et al. Ratiometric biosensors based on dimerization-dependent fluorescent protein exchange. Nat. Methods 12, 195–198 (2015).
Coleman, J. Structure and mechanism of alkaline phosphatase. Annu. Rev. Biophys. Biomol. Struct. 21, 441–483 (1992).
O'Brien, P.J. & Herschlag, D. Functional interrelationships in the alkaline phosphatase superfamily: phosphodiesterase activity of Escherichia coli alkaline phosphatase. Biochemistry 40, 5691–5699 (2001).
Alberstein, M., Eisenstein, M. & Abeliovich, H. Removing allosteric feedback inhibition of tomato 4-coumarate:CoA ligase by directed evolution. Plant J. 69, 57–69 (2012).
Yang, J.-S., Seo, S.W., Jang, S., Jung, G.Y. & Kim, S. Rational engineering of enzyme allosteric regulation through sequence evolution analysis. PLoS Comput. Biol. 8, e1002612 (2012).
Hu, X., Robin, S., O'Connell, S., Walsh, G. & Wall, J.G. Engineering of a fungal β-galactosidase to remove product inhibition by galactose. Appl. Microbiol. Biotechnol. 87, 1773–1782 (2010).
Coelho, P.S., Brustad, E.M., Kannan, A. & Arnold, F.H. Olefin cyclopropanation via carbene transfer catalyzed by engineered cytochrome P450 enzymes. Science 339, 307–310 (2013).
Pitsillides, C.M., Joe, E.K., Wei, X., Anderson, R.R. & Lin, C.P. Selective cell targeting with light-absorbing microparticles and nanoparticles. Biophys. J. 84, 4023–4032 (2003).
Wu, Y.-C. et al. Massively parallel delivery of large cargo into mammalian cells with light pulses. Nat. Methods 12, 439–444 (2015).
Otsu, N. A threshold selection method from gray-level histograms. IEEE SMC 9, 62–66 (1979).
Van Deventer, J.A. & Wittrup, K.D. Yeast surface display for antibody isolation: library construction, library screening, and affinity maturation. Methods Mol. Biol. 1131, 151–181 (2014).
Chao, G. et al. Isolating and engineering human antibodies using yeast surface display. Nat. Protoc. 1, 755–768 (2006).
Benton, D. & Krishnamoorthy, K. Performance of the parametric bootstrap method in small sample interval estimates. Adv. Appl. Stat. 2, 269–285 (2002).
Andrews, L.D., Zalatan, J.G. & Herschlag, D. Probing the origins of catalytic discrimination between phosphate and sulfate monoester hydrolysis: comparative analysis of alkaline phosphatase and protein tyrosine phosphatases. Biochemistry 53, 6811–6819 (2014).
Brune, M., Hunter, J.L., Corrie, J.E.T. & Webb, M.R. Direct, real-time measurement of rapid inorganic phosphate release using a novel fluorescent probe and its application to actomyosin subfragment 1 ATPase. Biochemistry 33, 8262–8271 (1994).
Zalatan, J.G., Fenn, T.D. & Herschlag, D. Comparative enzymology in the alkaline phosphatase superfamily to determine the catalytic role of an active-site metal ion. J. Mol. Biol. 384, 1174–1189 (2008).
Acknowledgements
The authors thank K.D. Wittrup and J. Van Deventer (MIT) and S. Sidhu (University of Toronto) for providing the yeast-displayed scFv library. This project was funded in part by the Stanford–Wallace H. Coulter Translational Partnership Award Program, the Siebel Stem Cell Institute and the Thomas and Stacey Siebel Foundation (to I.K.D.), the Stanford Photonics Research Center, a Hitachi America Faculty Scholar Award (to J.R.C.) and the US National Institutes of Health (GM49243 to D.H.). We acknowledge support from the National Science Foundation Graduate Fellowship Program (B.C. and A.K.), the Howard Hughes Medical Institute International Student Research Program (S.L.), a Fannie and John Hertz Foundation Graduate Fellowship (A.K.), the Stanford Bio-X Fellowship Program (S.L.), the Stanford Graduate Fellowship Program (B.C.) and the Stanford Dean's Fellowship Program (S.C.A.).
Author information
Authors and Affiliations
Contributions
All authors conceived and designed experiments and analyzed data. T.M.B., I.K.D. and B.C. designed and established the μSCALE platform. B.C., S.L. and I.K.D. established the μSCALE workflow. S.L. and B.C. performed protein engineering for binding interactions. S.C.A. and B.C. performed protein engineering for fluorescent proteins. A.K. and B.C. performed enzyme characterization and engineering with guidance from F.S. and D.H. The manuscript was prepared by B.C., S.L., A.K., S.C.A. and J.R.C. with input from all coauthors.
Corresponding authors
Ethics declarations
Competing interests
B.C., S.L., A.K., S.C.A, I.K.D, T.M.B. and J.R.C. are listed as inventors on patent applications related to technology described in this work. T.M.B. and I.K.D. are listed as inventors on pending patent applications (PCT/US2013/047792).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–18 and Supplementary Tables 1–7. (PDF 4020 kb)
Rights and permissions
About this article
Cite this article
Chen, B., Lim, S., Kannan, A. et al. High-throughput analysis and protein engineering using microcapillary arrays. Nat Chem Biol 12, 76–81 (2016). https://doi.org/10.1038/nchembio.1978
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.1978
This article is cited by
-
A high throughput bispecific antibody discovery pipeline
Communications Biology (2023)
-
Rapid discovery of high-affinity antibodies via massively parallel sequencing, ribosome display and affinity screening
Nature Biomedical Engineering (2023)
-
A biological camera that captures and stores images directly into DNA
Nature Communications (2023)
-
Single-cell sorting based on secreted products for functionally defined cell therapies
Microsystems & Nanoengineering (2022)
-
Directed evolution of Aspergillus oryzae lipase for the efficient resolution of (R,S)-ethyl-2-(4-hydroxyphenoxy) propanoate
Bioprocess and Biosystems Engineering (2020)