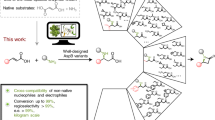
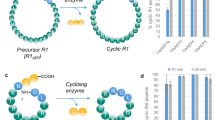
Abstract
Proteases are ubiquitous in nature, whereas naturally occurring peptide ligases, enzymes catalyzing the reverse reactions of proteases, are rare occurrences. Here we describe the discovery of butelase 1, to our knowledge the first asparagine/aspartate (Asx) peptide ligase to be reported. This highly efficient enzyme was isolated from Clitoria ternatea, a cyclic peptide–producing medicinal plant. Butelase 1 shares 71% sequence identity and the same catalytic triad with legumain proteases but does not hydrolyze the protease substrate of legumain. Instead, butelase 1 cyclizes various peptides of plant and animal origin with yields greater than 95%. With Kcat values of up to 17 s−1 and catalytic efficiencies as high as 542,000 M−1 s−1, butelase 1 is the fastest peptide ligase known. Notably, butelase 1 also displays broad specificity for the N-terminal amino acids of the peptide substrate, thus providing a new tool for C terminus–specific intermolecular peptide ligations.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Cascales, L. & Craik, D.J. Naturally occurring circular proteins: distribution, biosynthesis and evolution. Org. Biomol. Chem. 8, 5035–5047 (2010).
Craik, D.J. Chemistry—seamless proteins tie up their loose ends. Science 311, 1563–1564 (2006).
Wong, C.T. et al. Orally active peptidic bradykinin B1 receptor antagonists engineered from a cyclotide scaffold for inflammatory pain treatment. Angew. Chem. Int. Edn Engl. 51, 5620–5624 (2012).
Eisenbrandt, R. et al. Conjugative pili of IncP plasmids, and the Ti plasmid T pilus are composed of cyclic subunits. J. Biol. Chem. 274, 22548–22555 (1999).
Jack, R.W., Tagg, J.R. & Ray, B. Bacteriocins of gram-positive bacteria. Microbiol. Rev. 59, 171–200 (1995).
Sivonen, K., Leikoski, N., Fewer, D.P. & Jokela, J. Cyanobactins-ribosomal cyclic peptides produced by cyanobacteria. Appl. Microbiol. Biotechnol. 86, 1213–1225 (2010).
Tang, Y.Q. et al. A cyclic antimicrobial peptide produced in primate leukocytes by the ligation of two truncated α-defensins. Science 286, 498–502 (1999).
Cole, A.M. et al. Retrocyclin: a primate peptide that protects cells from infection by T- and M-tropic strains of HIV-1. Proc. Natl. Acad. Sci. USA 99, 1813–1818 (2002).
Luckett, S. et al. High-resolution structure of a potent, cyclic proteinase inhibitor from sunflower seeds. J. Mol. Biol. 290, 525–533 (1999).
Craik, D.J., Daly, N.L., Bond, T. & Waine, C. Plant cyclotides: a unique family of cyclic and knotted proteins that defines the cyclic cystine knot structural motif. J. Mol. Biol. 294, 1327–1336 (1999).
Arnison, P.G. et al. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 30, 108–160 (2013).
Trauger, J.W., Kohli, R.M., Mootz, H.D., Marahiel, M.A. & Walsh, C.T. Peptide cyclization catalysed by the thioesterase domain of tyrocidine synthetase. Nature 407, 215–218 (2000).
Sieber, S.A. & Marahiel, M.A. Learning from nature's drug factories: nonribosomal synthesis of macrocyclic peptides. J. Bacteriol. 185, 7036–7043 (2003).
Haase, J. & Lanka, E. A specific protease encoded by the conjugative DNA transfer systems of IncP and Ti plasmids is essential for pilus synthesis. J. Bacteriol. 179, 5728–5735 (1997).
Lee, J., McIntosh, J., Hathaway, B.J. & Schmidt, E.W. Using marine natural products to discover a protease that catalyzes peptide macrocyclization of diverse substrates. J. Am. Chem. Soc. 131, 2122–2124 (2009).
Barber, C.J. et al. The two-step biosynthesis of cyclic peptides from linear precursors in a member of the plant family Caryophyllaceae involves cyclization by a serine protease-like enzyme. J. Biol. Chem. 288, 12500–12510 (2013).
Craik, D.J. Host-defense activities of cyclotides. Toxins 4, 139–156 (2012).
Gruber, C.W. et al. Distribution and evolution of circular miniproteins in flowering plants. Plant Cell 20, 2471–2483 (2008).
Saska, I. et al. An asparaginyl endopeptidase mediates in vivo protein backbone cyclization. J. Biol. Chem. 282, 29721–29728 (2007).
Nguyen, G.K. et al. Discovery and characterization of novel cyclotides originated from chimeric precursors consisting of albumin-1 chain a and cyclotide domains in the Fabaceae family. J. Biol. Chem. 286, 24275–24287 (2011).
Poth, A.G. et al. Discovery of cyclotides in the Fabaceae plant family provides new insights into the cyclization, evolution, and distribution of circular proteins. ACS Chem. Biol. 6, 345–355 (2011).
Kembhavi, A.A., Buttle, D.J., Knight, C.G. & Barrett, A.J. The two cysteine endopeptidases of legume seeds: purification and characterization by use of specific fluorometric assays. Arch. Biochem. Biophys. 303, 208–213 (1993).
Sojka, D. et al. IrAE—an asparaginyl endopeptidase (legumain) in the gut of the hard tick Ixodes ricinus. Int. J. Parasitol. 37, 713–724 (2007).
Nguyen, G.K., Lim, W.H., Nguyen, P.Q. & Tam, J.P. Novel cyclotides and uncyclotides with highly shortened precursors from Chassalia chartacea and effects of methionine oxidation on bioactivities. J. Biol. Chem. 287, 17598–17607 (2012).
Conlan, B.F. et al. Insights into processing and cyclization events associated with biosynthesis of the cyclic peptide kalata B1. J. Biol. Chem. 287, 28037–28046 (2012).
Lee, J. & Bogyo, M. Development of near-infrared fluorophore (NIRF)-labeled activity-based probes for in vivo imaging of legumain. ACS Chem. Biol. 5, 233–243 (2010).
Becker, C. et al. Purification, cDNA cloning and characterization of proteinase-B, an asparagine-specific endopeptidase from germinating vetch (Vicia-sativa L) seeds. Eur. J. Biochem. 228, 456–462 (1995).
Abe, Y. et al. Asparaginyl endopeptidase of jack bean-seeds—purification, characterization, and high utility in protein-sequence analysis. J. Biol. Chem. 268, 3525–3529 (1993).
Dall, E. & Brandstetter, H. Mechanistic and structural studies on legumain explain its zymogenicity, distinct activation pathways, and regulation. Proc. Natl. Acad. Sci. USA 110, 10940–10945 (2013).
Hackeng, T.M., Griffin, J.H. & Dawson, P.E. Protein synthesis by native chemical ligation: expanded scope by using straightforward methodology. Proc. Natl. Acad. Sci. USA 96, 10068–10073 (1999).
Mao, H., Hart, S.A., Schink, A. & Pollok, B.A. Sortase-mediated protein ligation: a new method for protein engineering. J. Am. Chem. Soc. 126, 2670–2671 (2004).
Tam, J.P. & Wong, C.T. Chemical synthesis of circular proteins. J. Biol. Chem. 287, 27020–27025 (2012).
Tam, J.P., Lu, Y.A., Yang, J.L. & Chiu, K.W. An unusual structural motif of antimicrobial peptides containing end-to-end macrocycle and cystine-knot disulfides. Proc. Natl. Acad. Sci. USA 96, 8913–8918 (1999).
Kohli, R.M., Trauger, J.W., Schwarzer, D., Marahiel, M.A. & Walsh, C.T. Generality of peptide cyclization catalyzed by isolated thioesterase domains of nonribosomal peptide synthetases. Biochemistry 40, 7099–7108 (2001).
Ton-That, H., Liu, G., Mazmanian, S.K., Faull, K.F. & Schneewind, O. Purification and characterization of sortase, the transpeptidase that cleaves surface proteins of Staphylococcus aureus at the LPXTG motif. Proc. Natl. Acad. Sci. USA 96, 12424–12429 (1999).
Xu, M.Q. & Evans, T.C. Jr. Intein-mediated ligation and cyclization of expressed proteins. Methods. 24, 257–277 (2001).
Bolscher, J.G.M. et al. Sortase A as a tool for high-yield histatin cyclization. FASEB J. 25, 2650–2658 (2011).
Jia, X. et al. Semienzymatic cyclization of disulfide-rich peptides using sortase A. J. Biol. Chem. 289, 6627–6638 (2014).
Kimura, R.H., Tran, A.T. & Camarero, J.A. Biosynthesis of the cyclotide Kalata B1 by using protein splicing. Angew. Chem. Int. Edn Engl. 45, 973–976 (2006).
Austin, J., Kimura, R.H., Woo, Y.H. & Camarero, J.A. In vivo biosynthesis of an Ala-scan library based on the cyclic peptide SFTI-1. Amino Acids 38, 1313–1322 (2010).
Gould, A. et al. Recombinant production of rhesus theta-defensin-1 (RTD-1) using a bacterial expression system. Mol. Biosyst. 8, 1359–1365 (2012).
Austin, J., Wang, W., Puttamadappa, S., Shekhtman, A. & Camarero, J.A. Biosynthesis and biological screening of a genetically encoded library based on the cyclotide MCoTI-I. ChemBioChem 10, 2663–2670 (2009).
Young, T.S. et al. Evolution of cyclic peptide protease inhibitors. Proc. Natl. Acad. Sci. USA 108, 11052–11056 (2011).
Aboye, T.L. & Camarero, J.A. Biological synthesis of circular polypeptides. J. Biol. Chem. 287, 27026–27032 (2012).
Koehnke, J. et al. The mechanism of patellamide macrocyclization revealed by the characterization of the PatG macrocyclase domain. Nat. Struct. Mol. Biol. 19, 767–772 (2012).
Min, W. & Jones, D.H. In-vitro splicing of concanavalin-a is catalyzed by asparaginyl endopeptidase. Nat. Struct. Biol. 1, 502–504 (1994).
Gillon, A.D. et al. Biosynthesis of circular proteins in plants. Plant J. 53, 505–515 (2008).
Acknowledgements
We thank M. Bogyo at Stanford University for providing the legumain-specific probe LP-1 and R. Wang at Nanyang Technological University for helpful comments on this manuscript. This work was supported in part by the Singapore National Research Foundation grant NRF-CRP8-2011-05.
Author information
Authors and Affiliations
Contributions
G.K.T.N. designed the experiments and isolated and characterized butelase 1. S.W. performed the 1D NMR and homology modeling of butelase 1. Y.Q. synthesized the peptide libraries. X.H. evaluated the intermolecular ligation efficiency. Y.L. performed the kinetic studies for conotoxin, thanatin and histatin. J.P.T. initiated, planned, supervised and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Tables 1–3, Supplementary Figures 1–15 and Supplementary Note 1. (PDF 6225 kb)
Rights and permissions
About this article
Cite this article
Nguyen, G., Wang, S., Qiu, Y. et al. Butelase 1 is an Asx-specific ligase enabling peptide macrocyclization and synthesis. Nat Chem Biol 10, 732–738 (2014). https://doi.org/10.1038/nchembio.1586
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.1586
This article is cited by
-
Chemoenzymatic tandem cyclization for the facile synthesis of bicyclic peptides
Communications Chemistry (2024)
-
Recent advances in chemical protein synthesis: method developments and biological applications
Science China Chemistry (2024)
-
Self-cyclisation as a general and efficient platform for peptide and protein macrocyclisation
Communications Chemistry (2023)
-
Nature-inspired protein ligation and its applications
Nature Reviews Chemistry (2023)
-
C-terminal modification and functionalization of proteins via a self-cleavage tag triggered by a small molecule
Nature Communications (2023)