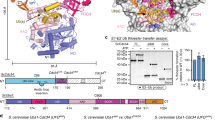
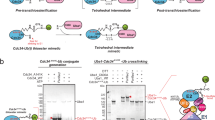
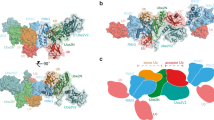
Abstract
Weak protein interactions between ubiquitin and the ubiquitin-proteasome system (UPS) enzymes that mediate its covalent attachment to substrates serve to position ubiquitin for optimal catalytic transfer. We show that a small-molecule inhibitor of the E2 ubiquitin-conjugating enzyme Cdc34A, called CC0651, acts by trapping a weak interaction between ubiquitin and the E2 donor ubiquitin-binding site. A structure of the ternary CC0651–Cdc34A–ubiquitin complex reveals that the inhibitor engages a composite binding pocket formed from Cdc34A and ubiquitin. CC0651 also suppresses the spontaneous hydrolysis rate of the Cdc34A-ubiquitin thioester without decreasing the interaction between Cdc34A and the RING domain subunit of the E3 enzyme. Stabilization of the numerous other weak interactions between ubiquitin and UPS enzymes by small molecules may be a feasible strategy to selectively inhibit different UPS activities.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Komander, D. & Rape, M. The ubiquitin code. Annu. Rev. Biochem. 81, 203–229 (2012).
Hoeller, D. & Dikic, I. Targeting the ubiquitin system in cancer therapy. Nature 458, 438–444 (2009).
Soucy, T.A., Dick, L.R., Smith, P.G., Milhollen, M.A. & Brownell, J.E. The NEDD8 conjugation pathway and its relevance in cancer biology and therapy. Genes Cancer 1, 708–716 (2010).
MacGurn, J.A., Hsu, P.C. & Emr, S.D. Ubiquitin and membrane protein turnover: from cradle to grave. Annu. Rev. Biochem. 81, 231–259 (2012).
Matyskiela, M.E. & Martin, A. Design principles of a universal protein degradation machine. J. Mol. Biol. 425, 199–213 (2013).
Ye, Y. & Rape, M. Building ubiquitin chains: E2 enzymes at work. Nat. Rev. Mol. Cell Biol. 10, 755–764 (2009).
Wenzel, D.M., Stoll, K.E. & Klevit, R.E. E2s: structurally economical and functionally replete. Biochem. J. 433, 31–42 (2011).
Saha, A., Lewis, S., Kleiger, G., Kuhlman, B. & Deshaies, R.J. Essential role for ubiquitin-ubiquitin-conjugating enzyme interaction in ubiquitin discharge from Cdc34 to substrate. Mol. Cell 42, 75–83 (2011).
Wickliffe, K.E., Lorenz, S., Wemmer, D.E., Kuriyan, J. & Rape, M. The mechanism of linkage-specific ubiquitin chain elongation by a single-subunit E2. Cell 144, 769–781 (2011).
Pruneda, J.N. et al. Structure of an E3:E2-Ub complex reveals an allosteric mechanism shared among RING/U-box ligases. Mol. Cell 47, 933–942 (2012).
Deshaies, R.J. & Joazeiro, C.A. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78, 399–434 (2009).
Emanuele, M.J. et al. Global identification of modular cullin-RING ligase substrates. Cell 147, 459–474 (2011).
Liao, H. et al. Quantitative proteomic analysis of cellular protein modulation upon inhibition of the NEDD8-activating enzyme by MLN4924. Mol. Cell Proteomics 10, M111.009183 (2011).
Kleiger, G., Saha, A., Lewis, S., Kuhlman, B. & Deshaies, R.J. Rapid E2–E3 assembly and disassembly enable processive ubiquitylation of cullin-RING ubiquitin ligase substrates. Cell 139, 957–968 (2009).
Duda, D.M. et al. Structure of a glomulin-RBX1–CUL1 complex: inhibition of a RING E3 ligase through masking of its E2-binding surface. Mol. Cell 47, 371–382 (2012).
Duda, D.M. et al. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell 134, 995–1006 (2008).
Soucy, T.A. et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 458, 732–736 (2009).
Brownell, J.E. et al. Substrate-assisted inhibition of ubiquitin-like protein-activating enzymes: the NEDD8 E1 inhibitor MLN4924 forms a NEDD8-AMP mimetic in situ. Mol. Cell 37, 102–111 (2010).
Ceccarelli, D.F. et al. An allosteric inhibitor of the human Cdc34 ubiquitin-conjugating enzyme. Cell 145, 1075–1087 (2011).
Spratt, D.E. & Shaw, G.S. Association of the disordered C-terminus of CDC34 with a catalytically bound ubiquitin. J. Mol. Biol. 407, 425–438 (2011).
Choi, Y.S. et al. The human Cdc34 carboxyl terminus contains a non-covalent ubiquitin binding activity that contributes to SCF-dependent ubiquitination. J. Biol. Chem. 285, 17754–17762 (2010).
Hamilton, K.S. et al. Structure of a conjugating enzyme-ubiquitin thiolester intermediate reveals a novel role for the ubiquitin tail. Structure 9, 897–904 (2001).
Plechanovová, A., Jaffray, E.G., Tatham, M.H., Naismith, J.H. & Hay, R.T. Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature 489, 115–120 (2012).
Dou, H., Buetow, L., Sibbet, G.J., Cameron, K. & Huang, D.T. BIRC7–E2 ubiquitin conjugate structure reveals the mechanism of ubiquitin transfer by a RING dimer. Nat. Struct. Mol. Biol. 19, 876–883 (2012).
Spratt, D.E., Wu, K., Kovacev, J., Pan, Z.Q. & Shaw, G.S. Selective recruitment of an E2–ubiquitin complex by an E3 ubiquitin ligase. J. Biol. Chem. 287, 17374–17385 (2012).
Capili, A.D. & Lima, C.D. Structure and analysis of a complex between SUMO and Ubc9 illustrates features of a conserved E2-Ubl interaction. J. Mol. Biol. 369, 608–618 (2007).
Knipscheer, P., van Dijk, W.J., Olsen, J.V., Mann, M. & Sixma, T.K. Noncovalent interaction between Ubc9 and SUMO promotes SUMO chain formation. EMBO J. 26, 2797–2807 (2007).
Seol, J.H. et al. Cdc53/cullin and the essential Hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme Cdc34. Genes Dev. 13, 1614–1626 (1999).
Saha, A. & Deshaies, R.J. Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol. Cell 32, 21–31 (2008).
Bracher, P.J., Snyder, P.W., Bohall, B.R. & Whitesides, G.M. The relative rates of thiol-thioester exchange and hydrolysis for alkyl and aryl thioalkanoates in water. Orig. Life Evol. Biosph. 41, 399–412 (2011).
Song, J. et al. Stability of thioester intermediates in ubiquitin-like modifications. Protein Sci. 18, 2492–2499 (2009).
Pierce, N.W., Kleiger, G., Shan, S.O. & Deshaies, R.J. Detection of sequential polyubiquitylation on a millisecond timescale. Nature 462, 615–619 (2009).
Burroughs, A.M., Jaffee, M., Iyer, L.M. & Aravind, L. Anatomy of the E2 ligase fold: implications for enzymology and evolution of ubiquitin/Ub-like protein conjugation. J. Struct. Biol. 162, 205–218 (2008).
Thiel, P., Kaiser, M. & Ottmann, C. Small-molecule stabilization of protein-protein interactions: an underestimated concept in drug discovery? Angew. Chem. Int. Edn Engl. 51, 2012–2018 (2012).
Schulman, B.A. & Harper, J.W. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nat. Rev. Mol. Cell Biol. 10, 319–331 (2009).
Pashkova, N. et al. WD40 repeat propellers define a ubiquitin-binding domain that regulates turnover of F box proteins. Mol. Cell 40, 433–443 (2010).
Reyes-Turcu, F.E., Ventii, K.H. & Wilkinson, K.D. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu. Rev. Biochem. 78, 363–397 (2009).
Husnjak, K. & Dikic, I. Ubiquitin-binding proteins: decoders of ubiquitin-mediated cellular functions. Annu. Rev. Biochem. 81, 291–322 (2012).
Ernst, A. et al. A strategy for modulation of enzymes in the ubiquitin system. Science 339, 590–595 (2013).
Zheng, N., Wang, P., Jeffrey, P.D. & Pavletich, N.P. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell 102, 533–539 (2000).
Tang, X. et al. Suprafacial orientation of the SCFCdc4 dimer accommodates multiple geometries for substrate ubiquitination. Cell 129, 1165–1176 (2007).
Delaglio, F. et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995).
Johnson, B.A. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol. Biol. 278, 313–352 (2004).
Wang, A.C., Grzesiek, S., Tschudin, R., Lodi, P.J. & Bax, A. Sequential backbone assignment of isotopically enriched proteins in D2O by deuterium-decoupled HA(CA)N and HA(CACO)N. J. Biomol. NMR 5, 376–382 (1995).
Fielding, L. NMR methods for the determination of protein–ligand dissociation constants. Prog. Nucl. Mag. Reson. Spectrosc. 51, 219–242 (2007).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
McCoy, A.J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Murshudov, G.N. et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 (2011).
Qian, X. et al. Discovery of the first potent and selective inhibitor of centromere-associated protein E: GSK923295. ACS Med. Chem. Lett. 1, 30–34 (2010).
Acknowledgements
We thank A. Ernst, R. Deshaies, A. van der Sloot, R. Grunberg, J.-F. Lavallée, C. James, K. Chan and F. Mercurio for helpful discussions. We also thank I. Kourinov and staff at the Argonne National Laboratory for assistance with microdiffraction experiments at the Advanced Photon Source on the Northeastern Collaborative Access Team beamlines, as supported by award RR-15301 from the National Center for Research Resources at the US National Institutes of Health (NIH) and contract DE-AC02-06CH11357 from the US Department of Energy. G.K. and A.Z. are funded by grants from the National Center for Research Resources (5P20RR016464-11) and the National Institute of General Medical Sciences (8 P20 GM103440-11) from the NIH. This work was supported by grants to F.S. and M.T. from the Canadian Institutes of Health Research (MOP-126129, MOP-57795); by an award from the Ministère de l'enseignement supérieur, de la recherche, de la science et de la technologie du Québec through Génome Québec to M.T.; by a Canadian Institutes of Health Research Postdoctoral Fellowship to D.J.S.-C.; by a Canada Research Chair in structural biology to F.S.; and by a Canada Research Chair in systems and synthetic biology to M.T.
Author information
Authors and Affiliations
Contributions
H.H. performed NMR; D.F.C. performed X-ray crystallography; S.O. and A.Z. performed ubiquitination assays; P.G. generated protein reagents; S.P. and D.J.S.-C. synthesized CC0651 and its analogs; M.A., S.S., A.M., G.K., M.T. and F.S. designed experiments and interpreted results; and M.T. and F.S. wrote the manuscript with contributions from all other authors.
Corresponding authors
Ethics declarations
Competing interests
A provisional patent application for E2 enzyme inhibition has been filed, with M.T. and F.S. as inventors.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Tables 1 and 2 and Supplementary Figures 1–12. (PDF 5931 kb)
Rights and permissions
About this article
Cite this article
Huang, H., Ceccarelli, D., Orlicky, S. et al. E2 enzyme inhibition by stabilization of a low-affinity interface with ubiquitin. Nat Chem Biol 10, 156–163 (2014). https://doi.org/10.1038/nchembio.1412
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.1412
This article is cited by
-
The ubiquitin system: orchestrating cellular signals in non-small-cell lung cancer
Cellular & Molecular Biology Letters (2020)
-
Unifying principles of bifunctional, proximity-inducing small molecules
Nature Chemical Biology (2020)
-
Structural insights into E1 recognition and the ubiquitin-conjugating activity of the E2 enzyme Cdc34
Nature Communications (2019)
-
Real-time tracking of complex ubiquitination cascades using a fluorescent confocal on-bead assay
BMC Biology (2018)
-
Differential proteomic analysis reveals sequential heat stress-responsive regulatory network in radish (Raphanus sativus L.) taproot
Planta (2018)