Abstract
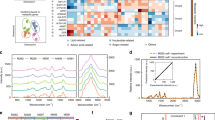
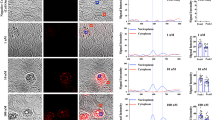
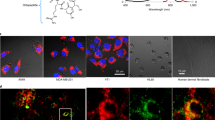
ABL1 tyrosine-kinase inhibitors (TKI) are front-line therapy for chronic myelogenous leukaemia and are among the best-known examples of targeted cancer therapeutics. However, the dynamic uptake into cells of TKIs of low molecular weight and their intracellular behaviour is unknown because of the difficulty of observing non-fluorescent small molecules at subcellular resolution. Here we report the direct label-free visualization and quantification of two TKI drugs (imatinib and nilotinib) inside living cells using hyperspectral stimulated Raman scattering imaging. Concentrations of both drugs were enriched over 1,000-fold in lysosomes as a result of their lysosomotropic properties. In addition, low solubility appeared to contribute significantly to the surprisingly large accumulation of nilotinib. We further show that the lysosomal trapping of imatinib was reduced more than tenfold when chloroquine is used simultaneously, which suggests that chloroquine may increase the efficacy of TKIs through lysosome-mediated drug–drug interaction in addition to the commonly proposed autophagy-inhibition mechanism.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Melo, J. V. & Barnes, D. J. Chronic myeloid leukaemia as a model of disease evolution in human cancer. Nature Rev. Cancer 7, 441–453 (2007).
Manley, P. W. et al. Structural resemblances and comparisons of the relative pharmacological properties of imatinib and nilotinib. Biorg. Med. Chem. 18, 6977–6986 (2010).
Adrian, F. J. et al. Allosteric inhibitors of Bcr-abl-dependent cell proliferation. Nature Chem. Biol. 2, 95–102 (2006).
Bellodi, C. et al. Targeting autophagy potentiates tyrosine kinase inhibitor-induced cell death in Philadelphia chromosome-positive cells, including primary CML stem cells. J. Clin. Invest. 119, 1109–1123 (2009).
Gupta, A. et al. Autophagy inhibition and antimalarials promote cell death in gastrointestinal stromal tumor (GIST). Proc. Natl Acad. Sci. 107, 14333–14338 (2010).
Lee, C. M. & Tannock, I. F. Inhibition of endosomal sequestration of basic anticancer drugs: influence on cytotoxicity and tissue penetration. Br. J. Cancer 94, 863–869 (2006).
Rubinsztein, D. C., Codogno, P. & Levine, B. Autophagy modulation as a potential therapeutic target for diverse diseases. Nature Rev. Drug Discov. 11, 709–730 (2012).
Verschooten, L. et al. Autophagy inhibitor chloroquine enhanced the cell death inducing effect of the flavonoid luteolin in metastatic squamous cell carcinoma cells. Plos One 7, e48264 (2012).
Lamoureux, F. et al. Blocked autophagy using lysosomotropic agents sensitizes resistant prostate tumor cells to the novel Akt inhibitor, AZD5363. Clin. Cancer. Res. 19, 833–844 (2013).
Zinn, R. et al. Combination treatment with ABT-737 and chloroquine in preclinical models of small cell lung cancer. Mol. Cancer 12, 16 (2013).
Maycotte, P. et al. Chloroquine sensitizes breast cancer cells to chemotherapy independent of autophagy. Autophagy 8, 200–212 (2012).
Satori, C. P. et al. Bioanalysis of eukaryotic organelles. Chem. Rev. 113, 2733–2811 (2013).
Baik, J. & Rosania, G. R. Molecular imaging of intracellular drug–membrane aggregate formation. Mol. Pharm. 8, 1742–1749 (2011).
Ling, J., Weitman, S. D., Miller, M. A., Moore, R. V. & Bovik, A. C. Direct Raman imaging techniques for study of the subcellular distribution of a drug. Appl. Opt. 41, 6006–6017 (2002).
Salehi, H. et al. Label-free detection of anticancer drug paclitaxel in living cells by confocal Raman microscopy. Appl. Phys. Lett. 102, 113701 (2013).
Matthäus, C. et al. in Confocal Raman Microscopy (eds Dieing, T., Hollricher, O. & Toporski, J.) Ch. 7, 137–163 (Springer Series in Optical Sciences 158, Springer, 2011).
Harada, Y. et al. Intracellular dynamics of topoisomerase I inhibitor, CPT-11, by slit-scanning confocal Raman microscopy. Histochem. Cell Biol. 132, 39–46 (2009).
Freudiger, C. W. et al. Label-free biomedical imaging with high sensitivity by stimulated Raman scattering microscopy. Science 322, 1857–1861 (2008).
Wang, M. C., Min, W., Freudiger, C. W., Ruvkun, G. & Xie, X. S. RNAi screening for fat regulatory genes with SRS microscopy. Nature Methods 8, 135–138 (2011).
Freudiger, C. W. et al. Multicolored stain-free histopathology with coherent Raman imaging. Lab. Invest. 92, 1492–1502 (2012).
Zhang, X. et al. Label-free live-cell imaging of nucleic acids using stimulated Raman scattering microscopy. Chemphyschem 13, 1054–1059 (2012).
Fu, D. et al. Quantitative chemical imaging with multiplex stimulated Raman scattering microscopy. J. Am. Chem. Soc. 134, 3623–3626 (2012).
Ozeki, Y. et al. High-speed molecular spectral imaging of tissue with stimulated Raman scattering. Nature Photon. 6, 845–851 (2012).
Fu, D., Holtom, G., Freudiger, C., Zhang, X. & Xie, X. S. Hyperspectral imaging with stimulated Raman scattering by chirped femtosecond lasers. J. Phys. Chem. B 117, 4634–4640 (2013).
Zhang, D. et al. Quantitative vibrational imaging by hyperspectral stimulated Raman scattering microscopy and multivariate curve resolution analysis. Anal. Chem. 85, 98–106 (2013).
Colthup, N. B., Daly, L. H. & Wiberley, S. E. Introduction to Infrared and Raman Spectroscopy (Academic Press, 1990).
Cînt[acaron]-Pînzaru, S. et al. FT-Raman and NIR-SERS characterization of the antimalarial drugs chloroquine and mefloquine and their interaction with hematin. J. Raman Spectrosc. 37, 326–334 (2006).
De Duve, C. et al. Lysosomotropic agents. Biochem. Pharmacol. 23, 2495–2531 (1974).
Trapp, S., Rosania, G. R., Horobin, R. W. & Kornhuber, J. Quantitative modeling of selective lysosomal targeting for drug design. Eur. Biophys. J. 37, 1317–1328 (2008).
Codogno, P. & Meijer, A. J. Autophagy and signaling: their role in cell survival and cell death. Cell Death Differ. 12, 1509–1518 (2005).
Kimura, T., Takabatake, Y., Takahashi, A. & Isaka, Y. Chloroquine in cancer therapy: a double-edged sword of autophagy. Cancer Res. 73, 3–7 (2013).
Poole, B. & Ohkuma, S. Effect of weak bases on the intralysosomal pH in mouse peritoneal macrophages. J. Cell Biol. 90, 665–669 (1981).
Zheng, N., Zhang, X. & Rosania, G. R. Effect of phospholipidosis on the cellular pharmacokinetics of chloroquine. J. Pharmacol. Exp. Ther. 336, 661–671 (2011).
Larson, R. A. et al. Imatinib pharmacokinetics and its correlation with response and safety in chronic-phase chronic myeloid leukemia: a subanalysis of the IRIS study. Blood 111, 4022–4028 (2008).
Deininger, M. W. Nilotinib . Clin. Cancer. Res. 14, 4027–4031 (2008).
Yamakoshi, H. et al. Alkyne-tag Raman imaging for visualization of mobile small molecules in live cells. J. Am. Chem. Soc. 134, 20681–20689 (2012).
Palacios, R. & Steinmetz, M. IL3-dependent mouse clones that express B-220 surface antigen, contain Ig genes in germ-line configuration, and generate B lymphocytes in vivo. Cell 41, 727–734 (1985).
Daley, G. Q. & Baltimore, D. Transformation of an interleukin 3-dependent hematopoietic cell line by the chronic myelogenous leukemia-specific P210bcr/abl protein. Proc. Natl Acad. Sci. 85, 9312–9316 (1988).
Acknowledgements
We thank S. Martin, J. Hastewell, M. Ji, F-K. Lu, C. Freudiger and W. Yang for helpful discussions. We also thank S. Moss for his help with the Raman peak assignments. This work was supported by the National Institute of Health's T-R01 (1R01EB010244-01) awarded to X.S.X.
Author information
Authors and Affiliations
Contributions
D.F., J.Z., Y.K.W. and X.S.X conceived the study. D.F. and J.Z. designed the study. D.F., J.Z., Y.K.W., X.S.X., and P.W.M participated extensively in the scientific discussion about the study. D.F. performed the hsSRS imaging study and analysed the imaging data. W.S.Z. performed the cell proliferation and phospho-STAT5 assay, with supervision from A.W., and J.Z., W.S.Z., A.W. and T.H. prepared the drug and cell samples. D.F., J.Z., P.M. and X.S.X. wrote the manuscript with contributions from W.S.Z., A.W. and T.H.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary information (PDF 619 kb)
Supplementary information
Supplementary Movie 1 (AVI 785 kb)
Supplementary information
Supplementary Movie 2 (AVI 785 kb)
Supplementary information
Supplementary Movie 3 (AVI 3204 kb)
Supplementary information
Supplementary Movie 4 (AVI 3204 kb)
Supplementary information
Supplementary Movie 5 (AVI 2348 kb)
Supplementary information
Supplementary Movie 6 (AVI 2348 kb)
Rights and permissions
About this article
Cite this article
Fu, D., Zhou, J., Zhu, W. et al. Imaging the intracellular distribution of tyrosine kinase inhibitors in living cells with quantitative hyperspectral stimulated Raman scattering. Nature Chem 6, 614–622 (2014). https://doi.org/10.1038/nchem.1961
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchem.1961
This article is cited by
-
Antidepressants enter cells, organelles, and membranes
Neuropsychopharmacology (2024)
-
Photoswitchable polyynes for multiplexed stimulated Raman scattering microscopy with reversible light control
Nature Communications (2024)
-
Fast Real-Time Brain Tumor Detection Based on Stimulated Raman Histology and Self-Supervised Deep Learning Model
Journal of Imaging Informatics in Medicine (2024)
-
Instant diagnosis of gastroscopic biopsy via deep-learned single-shot femtosecond stimulated Raman histology
Nature Communications (2022)
-
Flexible hyperspectral surface plasmon resonance microscopy
Nature Communications (2022)