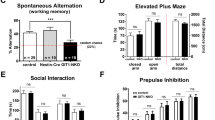
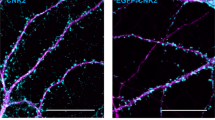
Abstract
Disrupted in schizophrenia 1 (DISC1), a genetic risk factor for multiple serious psychiatric diseases including schizophrenia, bipolar disorder and autism, is a key regulator of multiple neuronal functions linked to both normal development and disease processes. As these diseases are thought to share a common deficit in synaptic function and architecture, we have analyzed the role of DISC1 using an approach that focuses on understanding the protein–protein interactions of DISC1 specifically at synapses. We identify the Traf2 and Nck-interacting kinase (TNIK), an emerging risk factor itself for disease, as a key synaptic partner for DISC1, and provide evidence that the DISC1–TNIK interaction regulates synaptic composition and activity by stabilizing the levels of key postsynaptic density proteins. Understanding the novel DISC1–TNIK interaction is likely to provide insights into the etiology and underlying synaptic deficits found in major psychiatric diseases.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout









Similar content being viewed by others
References
Pardo CA, Eberhart CG . The neurobiology of autism. Brain Pathol 2007; 17: 434–447.
Sanacora G, Zarate CA, Krystal JH, Manji HK . Targeting the glutamatergic system to develop novel, improved therapeutics for mood disorders. Nat Rev Drug Discov 2008; 7: 426–437.
Sodhi M, Wood KH, Meador-Woodruff J . Role of glutamate in schizophrenia: integrating excitatory avenues of research. Expert Rev Neurother 2008; 8: 1389–1406.
Coyle JT . Glutamate and schizophrenia: beyond the dopamine hypothesis. Cell Mol Neurobiol 2006; 26: 365–384.
Schiffer HH . Glutamate receptor genes: susceptibility factors in schizophrenia and depressive disorders? Mol Neurobiol 2002; 25: 191–212.
Meador-Woodruff JH, Healy DJ . Glutamate receptor expression in schizophrenic brain. Brain Res Brain Res Rev 2000; 31: 288–294.
Ohnuma T, Kato H, Arai H, Faull RL, McKenna PJ, Emson PC . Gene expression of PSD95 in prefrontal cortex and hippocampus in schizophrenia. NeuroReport 2000; 11: 3133–3137.
Mirnics K, Middleton FA, Lewis DA, Levitt P . Analysis of complex brain disorders with gene expression microarrays: schizophrenia as a disease of the synapse. Trends Neurosci 2001; 24: 479–486.
Pennington K, Beasley CL, Dicker P, Fagan A, English J, Pariante CM et al. Prominent synaptic and metabolic abnormalities revealed by proteomic analysis of the dorsolateral prefrontal cortex in schizophrenia and bipolar disorder. Mol Psychiatry 2008; 13: 1102–1117.
Chubb JE, Bradshaw NJ, Soares DC, Porteous DJ, Millar JK . The DISC locus in psychiatric illness. Mol Psychiatry 2008; 13: 36–64.
Brandon NJ, Millar JK, Korth C, Sive H, Singh KK, Sawa A . Understanding the role of DISC1 in psychiatric disease and during normal development. J Neurosci 2009; 29: 12768–12775.
Millar JK, Wilson-Annan JC, Anderson S, Christie S, Taylor MS, Semple CA et al. Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum Mol Genet 2000; 9: 1415–1423.
St Clair D, Blackwood D, Muir W, Carothers A, Walker M, Spowart G et al. Association within a family of a balanced autosomal translocation with major mental illness. Lancet 1990; 336: 13–16.
Millar JK, Christie S, Anderson S, Lawson D, Hsiao-Wei Loh D, Devon RS et al. Genomic structure and localisation within a linkage hotspot of disrupted in schizophrenia 1, a gene disrupted by a translocation segregating with schizophrenia. Mol Psychiatry 2001; 6: 173–178.
Wang Q, Jaaro-Peled H, Sawa A, Brandon NJ . How has DISC1 enabled drug discovery? Mol Cell Neurosci 2008; 37: 187–195.
Millar JK, Pickard BS, Mackie S, James R, Christie S, Buchanan SR et al. DISC1 and PDE4B are interacting genetic factors in schizophrenia that regulate cAMP signaling. Science 2005; 310: 1187–1191.
Murdoch H, Mackie S, Collins DM, Hill EV, Bolger GB, Klussmann E et al. Isoform-selective susceptibility of DISC1/phosphodiesterase-4 complexes to dissociation by elevated intracellular cAMP levels. J Neurosci 2007; 27: 9513–9524.
Jaaro-Peled H, Hayashi-Takagi A, Seshadri S, Kamiya A, Brandon NJ, Sawa A . Neurodevelopmental mechanisms of schizophrenia: understanding disturbed postnatal brain maturation through neuregulin-1-ErbB4 and DISC1. Trends Neurosci 2009; 32: 485–495.
Kamiya A, Kubo K, Tomoda T, Takaki M, Youn R, Ozeki Y et al. A schizophrenia-associated mutation of DISC1 perturbs cerebral cortex development. Nat Cell Biol 2005; 7: 1167–1178.
Kim JY, Duan X, Liu CY, Jang MH, Guo JU, Pow-anpongkul N et al. DISC1 regulates new neuron development in the adult brain via modulation of AKT-mTOR signaling through KIAA1212. Neuron 2009; 63: 761–773.
Mao Y, Ge X, Frank CL, Madison JM, Koehler AN, Doud MK et al. Disrupted in schizophrenia 1 regulates neuronal progenitor proliferation via modulation of GSK3beta/beta-catenin signaling. Cell 2009; 136: 1017–1031.
Taya S, Shinoda T, Tsuboi D, Asaki J, Nagai K, Hikita T et al. DISC1 regulates the transport of the NUDEL/LIS1/14-3-3epsilon complex through kinesin-1. J Neurosci 2007; 27: 15–26.
Bradshaw NJ, Ogawa F, Antolin-Fontes B, Chubb JE, Carlyle BC, Christie S et al. DISC1, PDE4B, and NDE1 at the centrosome and synapse. Biochem Biophys Res Commun 2008; 377: 1091–1096.
Clapcote SJ, Lipina TV, Millar JK, Mackie S, Christie S, Ogawa F et al. Behavioral phenotypes of Disc1 missense mutations in mice. Neuron 2007; 54: 387–402.
Kirkpatrick B, Xu L, Cascella N, Ozeki Y, Sawa A, Roberts RC . DISC1 immunoreactivity at the light and ultrastructural level in the human neocortex. J Comp Neurol 2006; 497: 436–450.
Camargo LM, Collura V, Rain JC, Mizuguchi K, Hermjakob H, Kerrien S et al. Disrupted in schizophrenia 1 interactome: evidence for the close connectivity of risk genes and a potential synaptic basis for schizophrenia. Mol Psychiatry 2007; 12: 74–86.
Brandon NJ . Dissecting DISC1 function through protein-protein interactions. Biochem Soc Trans 2007; 35 (Part 5): 1283–1286.
Collins MO, Yu L, Coba MP, Husi H, Campuzano I, Blackstock WP et al. Proteomic analysis of in vivo phosphorylated synaptic proteins. J Biol Chem 2005; 280: 5972–5982.
Peng J, Kim MJ, Cheng D, Duong DM, Gygi SP, Sheng M . Semiquantitative proteomic analysis of rat forebrain postsynaptic density fractions by mass spectrometry. J Biol Chem 2004; 279: 21003–21011.
Fu CA, Shen M, Huang BC, Lasaga J, Payan DG, Luo Y . TNIK, a novel member of the germinal center kinase family that activates the c-Jun N-terminal kinase pathway and regulates the cytoskeleton. J Biol Chem 1999; 274: 30729–30737.
Taira K, Umikawa M, Takei K, Myagmar BE, Shinzato M, Machida N et al. The Traf2- and Nck-interacting kinase as a putative effector of Rap2 to regulate actin cytoskeleton. J Biol Chem 2004; 279: 49488–49496.
Mahmoudi T, Li VS, Ng SS, Taouatas N, Vries RG, Mohammed S et al. The kinase TNIK is an essential activator of Wnt target genes. EMBOJ 2009; 28: 3329–3340.
Glatt SJ, Everall IP, Kremen WS, Corbeil J, Sasik R, Khanlou N et al. Comparative gene expression analysis of blood and brain provides concurrent validation of SELENBP1 up-regulation in schizophrenia. Proc Natl Acad Sci USA 2005; 102: 15533–15538.
Matigian N, Windus L, Smith H, Filippich C, Pantelis C, McGrath J et al. Expression profiling in monozygotic twins discordant for bipolar disorder reveals dysregulation of the WNT signalling pathway. Mol Psychiatry 2007; 12: 815–825.
Potkin SG, Turner JA, Guffanti G, Lakatos A, Fallon JH, Nguyen DD et al. A genome-wide association study of schizophrenia using brain activation as a quantitative phenotype. Schizophr Bull 2009; 35: 96–108.
Shi J, Levinson DF, Duan J, Sanders AR, Zheng Y, Pe’er I et al. Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature 2009; 460: 753–757.
Banker GA, Cowan WM . Rat hippocampal neurons in dispersed cell culture. Brain Res 1977; 126: 397–425.
Kamiya A, Tomoda T, Chang J, Takaki M, Zhan C, Morita M et al. DISC1-NDEL1/NUDEL protein interaction, an essential component for neurite outgrowth, is modulated by genetic variations of DISC1. Hum Mol Genet 2006; 15: 3313–3323.
Hayashi-Takagi A, Takaki M, Graziane N, Seshadri S, Murdoch H, Dunlop AJ et al. Disrupted-in-schizophrenia 1 (DISC1) regulates spines of the glutamate synapse via Rac1. Nat Neurosci 2010; 13: 327–332.
Nonaka H, Takei K, Umikawa M, Oshiro M, Kuninaka K, Bayarjargal M et al. MINK is a Rap2 effector for phosphorylation of the postsynaptic scaffold protein TANC1. Biochem Biophys Res Commun 2008; 377: 573–578.
Brandon NJ, Schurov I, Camargo LM, Handford EJ, Duran-Jimeniz B, Hunt P et al. Subcellular targeting of DISC1 is dependent on a domain independent from the Nudel binding site. Mol Cell Neurosci 2005; 28: 613–624.
Shu H, Chen S, Bi Q, Mumby M, Brekken DL . Identification of phosphoproteins and their phosphorylation sites in the WEHI-231 B lymphoma cell line. Mol Cell Proteomics 2004; 3: 279–286.
Oppermann FS, Gnad F, Olsen JV, Hornberger R, Greff Z, Keri G et al. Large-scale proteomics analysis of the human kinome. Mol Cell Proteomics 2009; 8: 1751–1764.
Tomita S, Fukata M, Nicoll RA, Bredt DS . Dynamic interaction of stargazin-like TARPs with cycling AMPA receptors at synapses. Science 2004; 303: 1508–1511.
Chen L, Chetkovich DM, Petralia RS, Sweeney NT, Kawasaki Y, Wenthold RJ et al. Stargazin regulates synaptic targeting of AMPA receptors by two distinct mechanisms. Nature 2000; 408: 936–943.
Tomita S, Adesnik H, Sekiguchi M, Zhang W, Wada K, Howe JR et al. Stargazin modulates AMPA receptor gating and trafficking by distinct domains. Nature 2005; 435: 1052–1058.
Bats C, Groc L, Choquet D . The interaction between Stargazin and PSD-95 regulates AMPA receptor surface trafficking. Neuron 2007; 53: 719–734.
Colledge M, Snyder EM, Crozier RA, Soderling JA, Jin Y, Langeberg LK et al. Ubiquitination regulates PSD-95 degradation and AMPA receptor surface expression. Neuron 2003; 40: 595–607.
Ehlers MD . Reinsertion or degradation of AMPA receptors determined by activity-dependent endocytic sorting. Neuron 2000; 28: 511–525.
Newpher TM, Ehlers MD . Spine microdomains for postsynaptic signaling and plasticity. Trends Cell Biol 2009; 19: 218–227.
Kasai H, Fukuda M, Watanabe S, Hayashi-Takagi A, Noguchi J . Structural dynamics of dendritic spines in memory and cognition. Trends Neurosci 2010; 33: 121–129.
Penzes P, Jones KA . Dendritic spine dynamics—a key role for kalirin-7. Trends Neurosci 2008; 31: 419–427.
Collins DM, Murdoch H, Dunlop AJ, Charych E, Baillie GS, Wang Q et al. Ndel1 alters its conformation by sequestering cAMP-specific phosphodiesterase-4D3 (PDE4D3) in a manner that is dynamically regulated through protein kinase A (PKA). Cell Signal 2008; 20: 2356–2369.
Brandon NJ, Handford EJ, Schurov I, Rain JC, Pelling M, Duran-Jimeniz B et al. Disrupted in schizophrenia 1 and Nudel form a neurodevelopmentally regulated protein complex: implications for schizophrenia and other major neurological disorders. Mol Cell Neurosci 2004; 25: 42–55.
Enomoto A, Asai N, Namba T, Wang Y, Kato T, Tanaka M et al. Roles of disrupted-in-schizophrenia 1-interacting protein girdin in postnatal development of the dentate gyrus. Neuron 2009; 63: 774–787.
Shinoda T, Taya S, Tsuboi D, Hikita T, Matsuzawa R, Kuroda S et al. DISC1 regulates neurotrophin-induced axon elongation via interaction with Grb2. J Neurosci 2007; 27: 4–14.
Cingolani LA, Goda Y . Actin in action: the interplay between the actin cytoskeleton and synaptic efficacy. Nat Rev Neurosci 2008; 9: 344–356.
Acknowledgements
We thank Drs Carsten Korth, Hongjun Song and Atsushi Kamiya for reagents and scientific discussion. We thank Dr Pranab Chanda, Annette Sievers, Lora Cameron-Landis, Xiaotian Zhong and Adarsh Godbole for assistance in protein purification.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Molecular Psychiatry website
Supplementary information
Rights and permissions
About this article
Cite this article
Wang, Q., Charych, E., Pulito, V. et al. The psychiatric disease risk factors DISC1 and TNIK interact to regulate synapse composition and function. Mol Psychiatry 16, 1006–1023 (2011). https://doi.org/10.1038/mp.2010.87
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/mp.2010.87
Keywords
This article is cited by
-
Effects of MAP4K inhibition on neurite outgrowth
Molecular Brain (2023)
-
Rare coding variants as risk modifiers of the 22q11.2 deletion implicate postnatal cortical development in syndromic schizophrenia
Molecular Psychiatry (2023)
-
Structural interaction between DISC1 and ATF4 underlying transcriptional and synaptic dysregulation in an iPSC model of mental disorders
Molecular Psychiatry (2021)
-
TNIK influence the effects of antipsychotics on Wnt/β-catenin signaling pathway
Psychopharmacology (2021)
-
Human stem cell-based models for studying autism spectrum disorder-related neuronal dysfunction
Molecular Autism (2020)