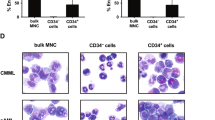
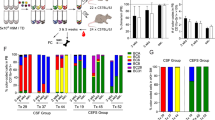
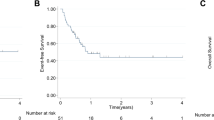
Abstract
Switches from the lymphoid to myeloid lineage during B-cell precursor acute lymphoblastic leukemia (BCP-ALL) treatment are considered rare and thus far have been detected in MLL-rearranged leukemia. Here, we describe a novel BCP-ALL subset, switching BCP-ALL or swALL, which demonstrated monocytosis early during treatment. Despite their monocytic phenotype, ‘monocytoids’ share immunoreceptor gene rearrangements with leukemic B lymphoblasts. All swALLs demonstrated BCP-ALL with CD2 positivity and no MLL alterations, and the proportion of swALLs cases among BCP-ALLs was unexpectedly high (4%). The upregulation of CEBPα and demethylation of the CEBPA gene were significant in blasts at diagnosis, prior to the time when most of the switching occurs. Intermediate stages between CD14negCD19posCD34pos B lymphoblasts and CD14posCD19negCD34neg ‘monocytoids’ were detected, and changes in the expression of PAX5, PU1, M-CSFR, GM-CSFR and other genes accompanied the switch. Alterations in the Ikaros and ERG genes were more frequent in swALL patients; however, both were altered in only a minority of swALLs. Moreover, switching could be recapitulated in vitro and in mouse xenografts. Although children with swALL respond slowly to initial therapy, risk-based ALL therapy appears the treatment of choice for swALL. SwALL shows that transdifferentiating into monocytic lineage is specifically associated with CEBPα changes and CD2 expression.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Stasik C, Ganguly S, Cunningham MT, Hagemeister S, Persons DL . Infant acute lymphoblastic leukemia with t(11;16)(q23;p13.3) and lineage switch into acute monoblastic leukemia. Cancer Genet Cytogenet 2006; 168: 146–149.
Derwich K, Sedek L, Meyer C, Pieczonka A, Dawidowska M, Gaworczyk A et al. Infant acute bilineal leukemia. Leuk Res. 2009; 33: 1005–1008.
Ridge SA, Cabrera ME, Ford AM, Tapia S, Risueno C, Labra S et al. Rapid intraclonal switch of lineage dominance in congenital leukaemia with a MLL gene rearrangement. Leukemia 1995; 9: 2023–2026.
Hutter C, Attarbaschi A, Fischer S, Meyer C, Dworzak M, Konig M et al. Acute monocytic leukaemia originating from MLL-MLLT3-positive pre-B cells. Br J Haematol 2010; 150: 621–623.
Rossi JG, Bernasconi AR, Alonso CN, Rubio PL, Gallego MS, Carrara CA et al. Lineage switch in childhood acute leukemia: an unusual event with poor outcome. Am J Hematol 2012; 87: 890–897.
Hrusak O, Porwit-MacDonald A . Antigen expression patterns reflecting genotype of acute leukemias. Leukemia 2002; 16: 1233–1258.
Marschalek R . Mechanisms of leukemogenesis by MLL fusion proteins. Br J Haematol 2011; 152: 141–154.
Oh SH, Park TS, Kim HR, Lee JY, Kim JH, Shin JH et al. Chronic myelogenous leukemia showing biphenotypic blast crisis followed by lineage switch to B lymphoblastic leukemia. Leuk Res 2009; 33: e195–e198.
Fronkova E, Mejstrikova E, Avigad S, Chik KW, Castillo L, Manor S et al. Minimal residual disease (MRD) analysis in the non-MRD-based ALL IC-BFM 2002 protocol for childhood ALL: is it possible to avoid MRD testing? Leukemia 2008; 22: 989–997.
Kawamoto H, Ikawa T, Masuda K, Wada H, Katsura Y . A map for lineage restriction of progenitors during hematopoiesis: the essence of the myeloid-based model. Immunol Rev 2010; 238: 23–36.
Montecino-Rodriguez E, Leathers H, Dorshkind K . Bipotential B-macrophage progenitors are present in adult bone marrow. Nat Immunol 2001; 2: 83–88.
Li J, Barreda DR, Zhang YA, Boshra H, Gelman AE, Lapatra S et al. B lymphocytes from early vertebrates have potent phagocytic and microbicidal abilities. Nat Immunol 2006; 7: 1116–1124.
Krejci O, Prouzova Z, Horvath O, Trka J, Hrusak O . Cutting edge: TCR delta gene is frequently rearranged in adult B lymphocytes. J Immunol 2003; 171: 524–527.
Fronkova E, Krejci O, Kalina T, Horvath O, Trka J, Hrusak O . Lymphoid differentiation pathways can be traced by TCR delta rearrangements. J Immunol 2005; 175: 2495–2500.
Mejstrikova E, Volejnikova J, Fronkova E, Zdrahalova K, Kalina T, Sterba J et al. Prognosis of children with mixed phenotype acute leukemia treated on the basis of consistent immunophenotypic criteria. Haematologica 2010; 95: 928–935.
Castro EC, Blazquez C, Boyd J, Correa H, de Chadarevian JP, Felgar RE et al. Clinicopathologic features of histiocytic lesions following ALL, with a review of the literature. Pediatr Dev Pathol 2010; 13: 225–237.
McClure R, Khoury J, Feldman A, Ketterling R . Clonal relationship between precursor B-cell acute lymphoblastic leukemia and histiocytic sarcoma: a case report and discussion in the context of similar cases. Leuk Res 2010; 34: e71–e73.
Congyang L, Xinggui W, Hao L, Weihua H . Synchronous histiocytic sarcoma and diffuse large B cell lymphoma involving the stomach: a case report and review of the literature. Int J Hematol 2011; 93: 247–252.
Feldman AL, Arber DA, Pittaluga S, Martinez A, Burke JS, Raffeld M et al. Clonally related follicular lymphomas and histiocytic/dendritic cell sarcomas: evidence for transdifferentiation of the follicular lymphoma clone. Blood 2008; 111: 5433–5439.
Bassarova A, Troen G, Fossa A, Ikonomou IM, Beiske K, Nesland JM et al. Transformation of B cell lymphoma to histiocytic sarcoma: somatic mutations of PAX-5 gene with loss of expression cannot explain transdifferentiation. J Hematop 2009; 2: 135–141.
Mejstrikova E, Kalina T, Trka J, Stary J, Hrusak O . Correlation of CD33 with poorer prognosis in childhood ALL implicates a potential of anti-CD33 frontline therapy. Leukemia 2005; 19: 1092–1094.
Mejstrikova E, Fronkova E, Kalina T, Omelka M, Batinic D, Dubravcic K et al. Detection of residual B precursor lymphoblastic leukemia by uniform gating flow cytometry. Pediatr Blood Cancer 2010; 54: 62–70.
Dworzak MN, Gaipa G, Ratei R, Veltroni M, Schumich A, Maglia O et al. Standardization of flow cytometric minimal residual disease evaluation in acute lymphoblastic leukemia: multicentric assessment is feasible. Cytometry B Clin Cytom 2008; 74: 331–340.
van der Velden VH, Cazzaniga G, Schrauder A, Hancock J, Bader P, Panzer-Grumayer ER et al. Analysis of minimal residual disease by Ig/TCR gene rearrangements: guidelines for interpretation of real-time quantitative PCR data. Leukemia 2007; 21: 604–611.
Zaliova M, Fronkova E, Krejcikova K, Muzikova K, Mejstrikova E, Stary J et al. Quantification of fusion transcript reveals a subgroup with distinct biological properties and predicts relapse in BCR/ABL-positive ALL: implications for residual disease monitoring. Leukemia 2009; 23: 944–951.
Petersen KB, Jusko WJ, Rasmussen M, Schmiegelow K . Population pharmacokinetics of prednisolone in children with acute lymphoblastic leukemia. Cancer Chemother Pharmacol 2003; 51: 465–473.
Gaipa G, Basso G, Aliprandi S, Migliavacca M, Vallinoto C, Maglia O et al. Prednisone induces immunophenotypic modulation of CD10 and CD34 in nonapoptotic B-cell precursor acute lymphoblastic leukemia cells. Cytometry B Clin Cytom 2008; 74: 150–155.
Andersen CL, Jensen JL, Orntoft TF . Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 2004; 64: 5245–5250.
Schmitz M, Breithaupt P, Scheidegger N, Cario G, Bonapace L, Meissner B et al. Xenografts of highly resistant leukemia recapitulate the clonal composition of the leukemogenic compartment. Blood 2011; 118: 1854–1864.
Mullighan CG, Su X, Zhang J, Radtke I, Phillips LA, Miller CB et al. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med 2009; 360: 470–480.
Radtke I, Mullighan CG, Ishii M, Su X, Cheng J, Ma J et al. Genomic analysis reveals few genetic alterations in pediatric acute myeloid leukemia. Proc Natl Acad Sci USA 2009; 106: 12944–12949.
Volejnikova J, Mejstrikova E, Dorge P, Meissner B, Zimmermannova O, Svojgr K et al. Ikaros (IKZF1) alterations and minimal residual disease at day 15 assessed by flow cytometry predict prognosis of childhood BCR/ABL-negative acute lymphoblastic leukemia. Pediatr Blood Cancer 2013; 60: 420–427.
Paz-Priel I, Friedman A . C/EBPalpha dysregulation in AML and ALL. Crit Rev Oncog 2011; 16: 93–102.
Schafer E, Irizarry R, Negi S, McIntyre E, Small D, Figueroa ME et al. Promoter hypermethylation in MLL-r infant acute lymphoblastic leukemia: biology and therapeutic targeting. Blood 2010; 115: 4798–4809.
Conter V, Bartram CR, Valsecchi MG, Schrauder A, Panzer-Grumayer R, Moricke A et al. Molecular response to treatment redefines all prognostic factors in children and adolescents with B-cell precursor acute lymphoblastic leukemia: results in 3184 patients of the AIEOP-BFM ALL 2000 study. Blood 2010; 115: 3206–3214.
Muhammad A, Schiller HB, Forster F, Eckerstorfer P, Geyeregger R, Leksa V et al. Sequential cooperation of CD2 and CD48 in the buildup of the early TCR signalosome. J Immunol 2009; 182: 7672–7680.
Uckun FM, Gaynon P, Sather H, Arthur D, Trigg M, Tubergen D et al. Clinical features and treatment outcome of children with biphenotypic CD2+ CD19+ acute lymphoblastic leukemia: a children's cancer group study. Blood 1997; 89: 2488–2493.
Yeoh EJ, Ross ME, Shurtleff SA, Williams WK, Patel D, Mahfouz R et al. Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell 2002; 1: 133–143.
Mullighan CG, Miller CB, Su XP, Radtke I, Dalton J, Song GC et al. ERG deletions define a novel subtype of B-progenitor acute lymphoblastic leukemia. ASH Annual Meeting Abstracts 2007; 110: 212A–213AA.
Clappier E, Auclerc M-F, Rapion J, Bakkus M, Caye A, Khemiri A et al. An intragenic ERG deletion is a marker of an oncogenic subtype of B-cell precursor acute lymphoblastic leukemia with a favorable outcome despite frequent IKZF1 deletions. Leukemia 2014; 28: 70–77.
Zaliova M, Zimmermanova O, Dorge P, Eckert C, Moricke A, Zimmermann M et al. ERG deletion is associated with CD2 and attenuates the negative impact of IKZF1 deletion in childhood acute lymphoblastic leukemia. Leukemia 2014; 28: 182–185.
Reynaud D, Demarco IA, Reddy KL, Schjerven H, Bertolino E, Chen Z et al. Regulation of B cell fate commitment and immunoglobulin heavy-chain gene rearrangements by Ikaros. Nat Immunol 2008; 9: 927–936.
Yin W, Rossin A, Clifford JL, Gronemeyer H . Co-resistance to retinoic acid and TRAIL by insertion mutagenesis into RAM. Oncogene 2006; 25: 3735–3744.
Xie H, Ye M, Feng R, Graf T . Stepwise reprogramming of B cells into macrophages. Cell 2004; 117: 663–676.
Di Tullio A, Manh TP, Schubert A, Mansson R, Graf T . CCAAT/enhancer binding protein {alpha} (C/EBP{alpha})-induced transdifferentiation of pre-B cells into macrophages involves no overt retrodifferentiation. Proc Natl Acad Sci USA 2011; 108: 17016–17021.
Laslo P, Spooner CJ, Warmflash A, Lancki DW, Lee HJ, Sciammas R et al. Multilineage transcriptional priming and determination of alternate hematopoietic cell fates. Cell 2006; 126: 755–766.
Yeamans C, Wang D, Paz-Priel I, Torbett BE, Tenen DG, Friedman AD . C/EBPalpha binds and activates the PU.1 distal enhancer to induce monocyte lineage commitment. Blood 2007; 110: 3136–3142.
Hohaus S, Petrovick MS, Voso MT, Sun Z, Zhang DE . Tenen DG. PU.1 (Spi-1) and C/EBP alpha regulate expression of the granulocyte-macrophage colony-stimulating factor receptor alpha gene. Mol Cell Biol 1995; 15: 5830–5845.
Akasaka T, Balasas T, Russell LJ, Sugimoto KJ, Majid A, Walewska R et al. Five members of the CEBP transcription factor family are targeted by recurrent IGH translocations in B-cell precursor acute lymphoblastic leukemia (BCP-ALL). Blood 2007; 109: 3451–3461.
Robinson HM, Taylor KE, Jalali GR, Cheung KL, Harrison CJ, Moorman AV . t(14;19)(q32;q13): a recurrent translocation in B-cell precursor acute lymphoblastic leukemia. Genes Chromosomes Cancer 2004; 39: 88–92.
Wouters BJ, Lowenberg B, Erpelinck-Verschueren CA, van Putten WL, Valk PJ, Delwel R . Double CEBPA mutations, but not single CEBPA mutations, define a subgroup of acute myeloid leukemia with a distinctive gene expression profile that is uniquely associated with a favorable outcome. Blood 2009; 113: 3088–3091.
Pieters R, Schrappe M, De Lorenzo P, Hann I, De Rossi G, Felice M et al. A treatment protocol for infants younger than 1 year with acute lymphoblastic leukaemia (Interfant-99): an observational study and a multicentre randomised trial. Lancet 2007; 370: 240–250.
Neale GA, Coustan-Smith E, Stow P, Pan Q, Chen X, Pui CH et al. Comparative analysis of flow cytometry and polymerase chain reaction for the detection of minimal residual disease in childhood acute lymphoblastic leukemia. Leukemia 2004; 18: 934–938.
Bierings M, Szczepanski T, van Wering ER, Willemse MJ, Langerak AW, Revesz T et al. Two consecutive immunophenotypic switches in a child with immunogenotypically stable acute leukaemia. Br J Haematol 2001; 113: 757–762.
Imataki O, Ohnishi H, Yamaoka G, Arai T, Kitanaka A, Kubota Y et al. Lineage switch from precursor B cell acute lymphoblastic leukemia to acute monocytic leukemia at relapse. Int J Clin Oncol 2010; 15: 112–115.
Acknowledgements
This work was supported by grants: Sciex 09.043 (LS), NT 12397-4 (EF), GAUK 914613 (LS), GACR P301/10/1877 and NT13462 (EM). The FACS Aria instrument was supported by EU-Prague project CZ.2.16/3.1.00/24022, FWvD was supported by the Kay Kendall Leukaemia Fund and ZZ was supported by RVO-VFN64165/2012. This work was also supported by the project for conceptual development of research organization 00064203 and CZ.2.16/3.1.00/24022.We thank Alzbeta Vazna for sequencing; Iveta Janotova for data management; Veronika Kanderova for performence of the in vitro experiments; Pavel Semerak for technical assistance in sorting; Daniel Thurner, Angela Schumich and Daniela Morf for processing of FC samples; Katerina Muzikova for PCR analyses of sorted samples; and the Czech Pediatric Hematology Group for collaboration (Doctors Sterba, Timr, Mihal, Cerna, Prochazkova, Blazek and Hak) and for providing clinical information (Doctors. Timr, Smisek, Votava and Hak).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Author Contributions
LS analyzed the 8CSAC combination and performed in vitro and in vivo experiments, methylation analyses and MRD examinations; JS designed the analysis of methylation status, and was involved in the design of in vitro experiments and their interpretation and designed the expression studies; EF was responsible for the MRD examinations and expression studies; EV evaluated the morphology; JV performed the MLPA; MZ, FWvD and JZ performed the SNP arrays; LR performed a part of the expression studies, ZZ was responsible for the cytogenetics; KP designed and performed the expression array, GC performed the expression studies; MF interpreted the methylation studies; TK designed the sorting; KF analyzed the profiling data; JPB designed the in vivo experiments and performed the investigations of the Swiss patient; BB executed the xenograft model; MND performed the investigations of the Austrian patients; JT supervised the molecular genetics; JS managed the patients and contributed to data collection; OH wrote and reviewed the manuscript and EM discovered the key aspects of the switching phenomenon, identified the swALL patients, designed the research, analyzed the data and wrote the manuscript. All authors have read and approved the submission of the manuscript.
Supplementary Information accompanies this paper on the Leukemia website
Supplementary information
Rights and permissions
About this article
Cite this article
Slamova, L., Starkova, J., Fronkova, E. et al. CD2-positive B-cell precursor acute lymphoblastic leukemia with an early switch to the monocytic lineage. Leukemia 28, 609–620 (2014). https://doi.org/10.1038/leu.2013.354
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2013.354
Keywords
This article is cited by
-
Integrated analysis of relapsed B-cell precursor Acute Lymphoblastic Leukemia identifies subtype-specific cytokine and metabolic signatures
Scientific Reports (2019)
-
Automated database-guided expert-supervised orientation for immunophenotypic diagnosis and classification of acute leukemia
Leukemia (2018)
-
CD19-negative relapse of pediatric B-cell precursor acute lymphoblastic leukemia following blinatumomab treatment
Blood Cancer Journal (2017)
-
The possible perils of targeted therapy
Leukemia (2016)
-
Independent development of lymphoid and histiocytic malignancies from a shared early precursor
Leukemia (2016)