Abstract
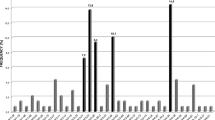
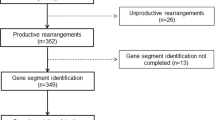
We performed an immunogenetic analysis of 345 IGHV–IGHD–IGHJ rearrangements from 337 cases with primary splenic small B-cell lymphomas of marginal-zone origin. Three immunoglobulin (IG) heavy variable (IGHV) genes accounted for 45.8% of the cases (IGHV1-2, 24.9%; IGHV4-34, 12.8%; IGHV3-23, 8.1%). Particularly for the IGHV1-2 gene, strong biases were evident regarding utilization of different alleles, with 79/86 rearrangements (92%) using allele *04. Among cases more stringently classified as splenic marginal-zone lymphoma (SMZL) thanks to the availability of splenic histopathological specimens, the frequency of IGHV1-2*04 peaked at 31%. The IGHV1-2*04 rearrangements carried significantly longer complementarity-determining region-3 (CDR3) than all other cases and showed biased IGHD gene usage, leading to CDR3s with common motifs. The great majority of analyzed rearrangements (299/345, 86.7%) carried IGHV genes with some impact of somatic hypermutation, from minimal to pronounced. Noticeably, 75/79 (95%) IGHV1-2*04 rearrangements were mutated; however, they mostly (56/75 cases; 74.6%) carried few mutations (97–99.9% germline identity) of conservative nature and restricted distribution. These distinctive features of the IG receptors indicate selection by (super)antigenic element(s) in the pathogenesis of SMZL. Furthermore, they raise the possibility that certain SMZL subtypes could derive from progenitor populations adapted to particular antigenic challenges through selection of VH domain specificities, in particular the IGHV1-2*04 allele.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Matutes E, Oscier D, Montalban C, Berger F, Callet-Bauchu E, Dogan A et al. Splenic marginal zone lymphoma proposals for a revision of diagnostic, staging and therapeutic criteria. Leukemia 2008; 22: 487–495.
Schmid C, Kirkham N, Diss T, Isaacson PG . Splenic marginal zone cell lymphoma. Am J Surg Pathol 1992; 16: 455–466.
Swerdlow S, Campo E, Harris NL et al. (eds) WHO Classification of Tumors of Haemopoietic and Lymphoid Tissues. IARC Press: Lyon, France, 2008.
Traverse-Glehen A, Baseggio L, Bauchu EC, Morel D, Gazzo S, Ffrench M et al. Splenic red pulp lymphoma with numerous basophilic villous lymphocytes: a distinct clinicopathologic and molecular entity? Blood 2008; 111: 2253–2260.
Matutes E . Immunophenotyping and differential diagnosis of hairy cell leukemia. Hematol Oncol Clin North Am 2006; 20: 1051–1063.
Stevenson FK, Sahota SS, Ottensmeier CH, Zhu D, Forconi F, Hamblin TJ . The occurrence and significance of V gene mutations in B cell-derived human malignancy. Adv Cancer Res 2001; 83: 81–116.
Dunn-Walters D, Thiede C, Alpen B, Spencer J . Somatic hypermutation and B-cell lymphoma. Philos Trans R Soc Lond B Biol Sci 2001; 356: 73–82.
Murray F, Darzentas N, Hadzidimitriou A, Tobin G, Boudjogra M, Scielzo C et al. Stereotyped patterns of somatic hypermutation in subsets of patients with chronic lymphocytic leukemia: implications for the role of antigen selection in leukemogenesis. Blood 2008; 111: 1524–1533.
Zhu D, Oscier DG, Stevenson FK . Splenic lymphoma with villous lymphocytes involves B cells with extensively mutated Ig heavy chain variable region genes. Blood 1995; 85: 1603–1607.
Algara P, Mateo MS, Sanchez-Beato M, Mollejo M, Navas IC, Romero L et al. Analysis of the IgV(H) somatic mutations in splenic marginal zone lymphoma defines a group of unmutated cases with frequent 7q deletion and adverse clinical course. Blood 2002; 99: 1299–1304.
Bahler DW, Pindzola JA, Swerdlow SH . Splenic marginal zone lymphomas appear to originate from different B cell types. Am J Pathol 2002; 161: 81–88.
Tierens A, Delabie J, Malecka A, Wang J, Gruszka-Westwood A, Catovsky D et al. Splenic marginal zone lymphoma with villous lymphocytes shows on-going immunoglobulin gene mutations. Am J Pathol 2003; 162: 681–689.
Papadaki T, Stamatopoulos K, Belessi C, Pouliou E, Parasi A, Douka V et al. Splenic marginal-zone lymphoma: one or more entities? A histologic, immunohistochemical, and molecular study of 42 cases. Am J Surg Pathol 2007; 31: 438–446.
Stamatopoulos K, Belessi C, Papadaki T, Kalagiakou E, Stavroyianni N, Douka V et al. Immunoglobulin heavy- and light-chain repertoire in splenic marginal zone lymphoma. Mol Med 2004; 10: 89–95.
Hockley SL, Giannouli S, Morilla A, Wotherspoon A, Morgan GJ, Matutes E et al. Insight into the molecular pathogenesis of hairy cell leukaemia, hairy cell leukaemia variant and splenic marginal zone lymphoma, provided by the analysis of their IGH rearrangements and somatic hypermutation patterns. Br J Haematol 2009; 148: 666–669.
Traverse-Glehen A, Davi F, Ben Simon E, Callet-Bauchu E, Felman P, Baseggio L et al. Analysis of VH genes in marginal zone lymphoma reveals marked heterogeneity between splenic and nodal tumors and suggests the existence of clonal selection. Haematologica 2005; 90: 470–478.
Dunn-Walters DK, Boursier L, Spencer J, Isaacson PG . Analysis of immunoglobulin genes in splenic marginal zone lymphoma suggests ongoing mutation. Hum Pathol 1998; 29: 585–593.
Zhu D, Orchard J, Oscier DG, Wright DH, Stevenson FK . V(H) gene analysis of splenic marginal zone lymphomas reveals diversity in mutational status and initiation of somatic mutation in vivo. Blood 2002; 100: 2659–2661.
Arcaini L, Zibellini S, Passamonti F, Rattotti S, Lucioni M, Invernizzi R et al. Splenic marginal zone lymphoma: clinical clustering of immunoglobulin heavy chain repertoires. Blood Cells Mol Dis 2009; 42: 286–291.
Rinaldi A, Forconi F, Arcaini L, Mian M, Sozzi E, Zibellini S et al. Immunogenetics features and genomic lesions in splenic marginal zone lymphoma. Br J Haematol 2010; 151: 435–439.
Zibellini S, Capello D, Forconi F, Marcatili P, Rossi D, Rattotti S et al. Stereotyped patterns of B-cell receptor in splenic marginal zone lymphoma. Haematologica 2010; 95: 1792–1796.
Salido M, Baro C, Oscier D, Stamatopoulos K, Dierlamm J, Matutes E et al. Cytogenetic aberrations and their prognostic value in a series of 330 splenic marginal zone B-cell lymphomas: a multicenter study of the Splenic B-Cell Lymphoma Group. Blood 2010; 116: 1479–1488.
Tracey L, Aggarwal M, Garcia-Cosio M, Villuendas R, Algara P, Sanchez-Beato M et al. Somatic hypermutation signature in B-cell low-grade lymphomas. Haematologica 2008; 93: 1186–1194.
Sutton LA, Kostareli E, Hadzidimitriou A, Darzentas N, Tsaftaris A, Anagnostopoulos A et al. Extensive intraclonal diversification in a subgroup of chronic lymphocytic leukemia patients with stereotyped IGHV4-34 receptors: implications for ongoing interactions with antigen. Blood 2009; 114: 4460–4468.
Giudicelli V, Duroux P, Ginestoux C, Folch G, Jabado-Michaloud J, Chaume D et al. IMGT/LIGM-DB, the IMGT comprehensive database of immunoglobulin and T cell receptor nucleotide sequences. Nucleic Acids Res 2006; 34 (Database issue): D781–D784.
Lefranc MP, Giudicelli V, Ginestoux C, Jabado-Michaloud J, Folch G, Bellahcene F et al. IMGT, the international ImMunoGeneTics information system. Nucleic Acids Res 2009; 37 (Database issue): D1006–D1012.
Brochet X, Lefranc MP, Giudicelli V . IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V–J and V–D–J sequence analysis. Nucleic Acids Res 2008; 36: W503–W508.
Darzentas N, Hadzidimitriou A, Murray F, Hatzi K, Josefsson P, Laoutaris N et al. A different ontogenesis for chronic lymphocytic leukemia cases carrying stereotyped antigen receptors: molecular and computational evidence. Leukemia 2009; 24: 125–132.
Emsley P, Lohkamp B, Scott WG, Cowtan K . Features and development of Coot. Acta Crystallogr D Biol Crystallogr 2010; 66 (Part 4): 486–501.
Gordon JC, Myers JB, Folta T, Shoja V, Heath LS, Onufriev A . H++: a server for estimating pKas and adding missing hydrogens to macromolecules. Nucleic Acids Res 2005; 33 (Web Server issue): W368–W371.
Hess B . GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput 2008; 4: 435–447.
DePaul AJ, Thompson EJ, Patel SS, Haldeman K, Sorin EJ . Equilibrium conformational dynamics in an RNA tetraloop from massively parallel molecular dynamics. Nucleic Acids Res 2010; 38: 4856–4867.
Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C . Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins 2006; 65: 712–725.
Sorin EJ, Pande VS . Exploring the helix–coil transition via all-atom equilibrium ensemble simulations. Biophys J 2005; 88: 2472–2493.
Stanfield RL, Takimoto-Kamimura M, Rini JM, Profy AT, Wilson IA . Major antigen-induced domain rearrangements in an antibody. Structure 1993; 1: 83–93.
Langerak A, Davi F, Ghia P, Hadzidimitriou A, Murray F, Potter K et al. Immunoglobulin sequence analysis and prognostication in CLL: guidelines from the ERIC review board for reliable interpretation of problematic cases. Leukemia 2011; 25: 979–984.
Dunn-Walters DK, Isaacson PG, Spencer J . Analysis of mutations in immunoglobulin heavy chain variable region genes of microdissected marginal zone (MGZ) B cells suggests that the MGZ of human spleen is a reservoir of memory B cells. J Exp Med 1995; 182: 559–566.
Stein K, Hummel M, Korbjuhn P, Foss HD, Anagnostopoulos I, Marafioti T et al. Monocytoid B cells are distinct from splenic marginal zone cells and commonly derive from unmutated naive B cells and less frequently from postgerminal center B cells by polyclonal transformation. Blood 1999; 94: 2800–2808.
Rassenti LZ, Kipps TJ . Lack of allelic exclusion in B cell chronic lymphocytic leukemia. J Exp Med 1997; 185: 1435–1445.
Belessi C, Stamatopoulos K, Hadzidimitriou A, Hatzi K, Smilevska T, Stavroyianni N et al. Analysis of expressed and non-expressed IGK locus rearrangements in chronic lymphocytic leukemia. Mol Med 2005; 11: 52–58.
Hadzidimitriou A, Darzentas N, Murray F, Smilevska T, Arvaniti E, Tresoldi C et al. Evidence for the significant role of immunoglobulin light chains in antigen recognition and selection in chronic lymphocytic leukemia. Blood 2009; 113: 403–411.
Kenny JJ, Rezanka LJ, Lustig A, Fischer RT, Yoder J, Marshall S et al. Autoreactive B cells escape clonal deletion by expressing multiple antigen receptors. J Immunol 2000; 164: 4111–4119.
Li Y, Li H, Weigert M . Autoreactive B cells in the marginal zone that express dual receptors. J Exp Med 2002; 195: 181–188.
Chiorazzi N, Ferrarini M . Cellular origin(s) of chronic lymphocytic leukemia: cautionary notes and additional considerations and possibilities. Blood 2011; 117: 1781–1791.
Barbas SM, Ditzel HJ, Salonen EM, Yang WP, Silverman GJ, Burton DR . Human autoantibody recognition of DNA. Proc Natl Acad Sci USA 1995; 92: 2529–2533.
Rahman A, Giles I, Haley J, Isenberg D . Systematic analysis of sequences of anti-DNA antibodies—relevance to theories of origin and pathogenicity. Lupus 2002; 11: 807–823.
Pommie C, Levadoux S, Sabatier R, Lefranc G, Lefranc MP . IMGT standardized criteria for statistical analysis of immunoglobulin V-REGION amino acid properties. J Mol Recognit 2004; 17: 17–32.
Xu JL, Davis MM . Diversity in the CDR3 region of V(H) is sufficient for most antibody specificities. Immunity 2000; 13: 37–45.
Jang YJ, Stollar BD . Anti-DNA antibodies: aspects of structure and pathogenicity. Cell Mol Life Sci 2003; 60: 309–320.
Weitkamp JH, Kallewaard NL, Bowen AL, Lafleur BJ, Greenberg HB, Crowe Jr JE . VH1-46 is the dominant immunoglobulin heavy chain gene segment in rotavirus-specific memory B cells expressing the intestinal homing receptor alpha4beta7. J Immunol 2005; 174: 3454–3460.
Barrios Y, Jirholt P, Ohlin M . Length of the antibody heavy chain complementarity determining region 3 as a specificity-determining factor. J Mol Recognit 2004; 17: 332–338.
Silberstein LE, George A, Durdik JM, Kipps TJ . The V4-34 encoded anti-i autoantibodies recognize a large subset of human and mouse B-cells. Blood Cells Mol Dis 1996; 22: 126–138.
Kruetzmann S, Rosado MM, Weber H, Germing U, Tournilhac O, Peter HH et al. Human immunoglobulin M memory B cells controlling Streptococcus pneumoniae infections are generated in the spleen. J Exp Med 2003; 197: 939–945.
Seifert M, Kuppers R . Molecular footprints of a germinal center derivation of human IgM+(IgD+)CD27+ B cells and the dynamics of memory B cell generation. J Exp Med 2009; 206: 2659–2669.
Weill JC, Weller S, Reynaud CA . Human marginal zone B cells. Annu Rev Immunol 2009; 27: 267–285.
Acknowledgements
We express our gratitude to Professor Marie-Paule Lefranc (Laboratoire d’Immunogenetique Moleculaire, LIGM, Universite Montpellier II, Montpellier, France) for valuable support, suggestions and guidance in immunoglobulin gene sequence analysis. The analysis of French cases was supported by grants from the Comité du Rhône de la Ligue Nationale contre le Cancer. The analysis of the Spanish cases was supported by grants from the AECC and the Ministerio de Ciencia e Innovación, Spain (SAF2008-03871, RETICS). The analysis of cases from the Institute of Cancer Research (London, UK) was supported by the UK Cancer Research Fund. The antibody molecular dynamics simulation was supported by a research grant from the Italian Ministry for University and Research (FIRB) and the CARIPLO Foundation, Milano, Italy.
Author Contributions
Vasilis Bikos performed research, analyzed data and wrote the paper. Nikos Darzentas and Anastasia Hadzidimitriou performed research and analyzed data. Zadie Davis, Sarah Hockley, Alexandra Traverse-Glehen, Patricia Algara, Alessandra Santoro, David Gonzalez, Manuela Mollejo, Antonis Dagklis and Pascale Felman performed research. George Bourikas, Achilles Anagnostopoulos and Athanasios Tsaftaris supervised research. Fabrizio Gangemi and Massimo Degano performed the antibody molecular dynamics simulation. Emilio Iannitto, Maurilio Ponzoni, Francoise Berger, Chrysoula Belessi, Paolo Ghia, Theodora Papadaki, Ahmet Dogan, Estella Matutes, Miguel Angel Piris and David Oscier provided samples and associated clinicopathological data, and supervised research. Kostas Stamatopoulos designed the study, supervised research and wrote the paper.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Leukemia website
Supplementary information
Rights and permissions
About this article
Cite this article
Bikos, V., Darzentas, N., Hadzidimitriou, A. et al. Over 30% of patients with splenic marginal zone lymphoma express the same immunoglobulin heavy variable gene: ontogenetic implications. Leukemia 26, 1638–1646 (2012). https://doi.org/10.1038/leu.2012.3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2012.3
Keywords
This article is cited by
-
Tracing CLL-biased stereotyped immunoglobulin gene rearrangements in normal B cell subsets using a high-throughput immunogenetic approach
Molecular Medicine (2020)
-
New roles for B cell receptor associated kinases: when the B cell is not the target
Leukemia (2019)
-
Lymphoma-like monoclonal B cell lymphocytosis in a patient population: biology, natural evolution, and differences from CLL-like clones
Annals of Hematology (2018)
-
Antigen receptor stereotypy in chronic lymphocytic leukemia
Leukemia (2017)
-
Clinical utility of recently identified diagnostic, prognostic, and predictive molecular biomarkers in mature B-cell neoplasms
Modern Pathology (2017)