Abstract
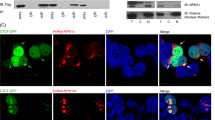
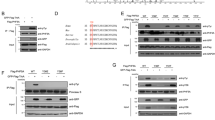
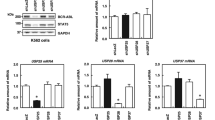
PTEN (phosphatase and tensin homolog deleted in chromosome 10) is a bona fide dual lipid and protein phosphatase with cytoplasmic (Cy) and nuclear localization. PTEN nuclear exclusion has been associated with tumorigenesis. Nucleophosmin (NPM1) is frequently mutated in acute myeloid leukemia (AML) and displays Cy localization in mutated nucleophosmin (NPMc+) AML. Here we show that NPM1 directly interacts with herpes virus-associated ubiquitin specific protease (HAUSP), which is known as a PTEN deubiquitinating enzyme. Strikingly, PTEN is aberrantly localized in AML carrying NPMc+. Mechanistically, NPM1 in the nucleus opposes HAUSP-mediated deubiquitination and this promotes the shuttle of PTEN to the cytoplasm. In the cytoplasm, NPMc+ prevents HAUSP from deubiquitinating PTEN, causing the latter to stay in the cytoplasm where it is polyubiquitinated and degraded. Our findings delineate a new NPM1–HAUSP molecular interaction controlling PTEN deubiquitination and trafficking.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Song MS, Salmena L, Pandolfi PP . The functions and regulation of the PTEN tumour suppressor. Nat Rev Mol Cell Biol 2012; 13: 283–296.
Chung JH, Eng C . Nuclear-cytoplasmic partitioning of phosphatase and tensin homologue deleted on chromosome 10 (PTEN) differentially regulates the cell cycle and apoptosis. Cancer Res 2005; 65: 8096–8100.
Li DM, Sun H . TEP1, encoded by a candidate tumor suppressor locus, is a novel protein tyrosine phosphatase regulated by transforming growth factor beta. Cancer Res 1997; 57: 2124–2129.
Planchon SM, Waite KA, Eng C . The nuclear affairs of PTEN. J Cell Sci 2008; 121: 249–253.
Shen WH, Balajee AS, Wang J, Wu H, Eng C, Pandolfi PP et al. Essential role for nuclear PTEN in maintaining chromosomal integrity. Cell 2007; 128: 157–170.
Hollander MC, Blumenthal GM, Dennis PA . PTEN loss in the continuum of common cancers, rare syndromes and mouse models. Nature Rev Cancer 2011; 11: 289–301.
Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science 1997; 275: 1943–1947.
Peterson LM, Kipp BR, Halling Kerr SE, Smith DI, Distad TJ et al. Molecular characterization of endometrial cancer: a correlative study assessing microsatellite instability, MLH1 hypermethylation, DNA mismatch repair protein expression, and PTEN, PIK3CA, KRAS, and BRAF mutation analysis. Int J Gynecol Pathol 2012; 31: 195–205.
Steck PA, Pershouse MA, Jasser SA, Yung WK, Lin H, Ligon AH et al. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat Genet 1997; 15: 356–362.
Patel JP, Gönen M, Figueroa ME, Fernandez H, Sun Z, Racevskis J et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med 2012; 366: 1079–1089.
Aggerholm A, Grønbaek K, Guldberg P, Hokland P . Mutational analysis of the tumour suppressor gene MMAC1/PTEN in malignant myeloid disorders. Eur J Haematol 2000; 65: 109–113.
Liu TC, Lin PM, Chang JG, Lee JP, Chen TP, Lin SF . Mutation analysis of PTEN/MMAC1 in acute myeloid leukaemia. Am J Hematol 2000; 63: 170–175.
Fridberg M, Servin A, Anagnostaki L, Linderoth J, Berglund M, Söderberg O et al. Protein expression and cellular localization in two prognostic subgroups of diffuse large B-cell lymphoma: higher expression of ZAP70 and PKC-beta II in the non-germinal center group and poor survival in patients deficient in nuclear PTEN. Leuk Lymphoma 2007; 48: 2221–2232.
Yin Y, Shen WH . PTEN: a new guardian of the genome. Oncogene 2008; 27: 5443–5453.
Song MS, Salmena L, Carracedo A, Egia A, Lo-Coco F, Teruya-Feldstein J et al. The deubiquitinylation and localization of PTEN are regulated by a HAUSP-PML network. Nature 2008; 455: 813–817.
Trotman LC, Wang X, Alimonti A, Chen Z, Teruya-Feldstein J, Yang H et al. Ubiquitination regulates PTEN nuclear import and tumor suppression. Cell 2007; 128: 141–156.
Mund T, Pelham HR . Regulation of PTEN/Akt and MAP kinase signaling pathways by the ubiquitin ligase activators Ndfip1 and Ndfip2. Proc Natl Acad Sci USA 2010; 107: 11429–11434.
Van Themsche C, Leblanc V, Parent S, Asselin E . X-linked inhibitor of apoptosis protein (XIAP) regulates PTEN ubiquitination, content, and compartmentalization. J Biol Chem 2009; 284: 20462–20466.
Wang X, Trotman LC, Koppie T, Alimonti A, Chen Z, Gao Z et al. NEDD4-1 is a proto-oncogenic ubiquitin ligase for PTEN. 2007 Cell 128: 129–139.
Falini B, Mecucci C, Tiacci E, Alcalay M, Rosati R, Pasqualucci L et al. GIMEMA Acute Leukemia Working Party. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med 2005; 352: 254–266.
Falini B, Bolli N, Shan J, Martelli MP, Liso A, Pucciarini A et al. Both carboxy-terminus NES motif and mutated tryptophan(s) are crucial for aberrant nuclear export of nucleophosmin leukemic mutants in NPMc+ AML. Blood 2006; 10: 4514–4523.
Colombo E, Martinelli P, Zamponi R, Shing DC, Bonetti P, Luzi L et al. Delocalization and destabilization of the Arf tumor suppressor by the leukaemia-associated NPM mutant. Cancer Res 2006; 66: 3044–3050.
Grisendi S, Mecucci C, Falini B, Pandolfi PP . Nucleophosmin and cancer. Nat Rev Cancer 2006; 6: 493–505.
van Dongen JJ, Macintyre EA, Gabert JA, Delabesse E, Rossi V, Saglio G et al. Standardized RT-PCR analysis of fusion gene transcripts from chromosome aberrations in acute leukemia for detection of minimal residual disease. Report of the BIOMED-1 Concerted Action: investigation of minimal residual disease in acute leukemia. Leukemia 1999; 13: 1901–1928.
Noguera NI, Ammatuna E, Zangrilli D, Lavorgna S, Divona M, Buccisano F et al. Simultaneous detection of NPM1 and FLT3-ITD mutations by capillary electrophoresis in acute myeloid leukemia. Leukemia 2005; 19: 1479–1482.
Noguera NI, Breccia M, Divona M, Diverio D, Costa V, De Santis S et al. Alterations of the FLT3 gene in acute promyelocytic leukemia: association with diagnostic characteristics and analysis of clinical outcome in patients treated with the Italian AIDA protocol. Leukemia 2002; 16: 2185–2189.
Gruszka AM, Lavorgna S, Consalvo MI, Ottone T, Martinelli C, Cinquanta M et al. A monoclonal antibody against mutated nucleophosmin 1 for the molecular diagnosis of acute myeloid leukemias. Blood 2010; 116: 2096–2102.
Anderson DH . p85 plays a critical role in controlling flux through the PI3K/PTEN signaling axis through dual regulation of both p110 (PI3K) and PTEN. Cell Cycle 2010; 9: 2055–2056.
Chagpar RB, Links PH, Pastor MC, Furber LA, Hawrysh AD, Chamberlain MD et al. Direct positive regulation of PTEN by the p85 subunit of phosphatidylinositol 3-kinase. Proc Natl Acad Sci USA 2010; 107: 5471–5476.
Rabinovsky R, Pochanard P, McNear C, Brachmann SM, Duke-Cohan JS, Garraway LA et al. p85 Associates with unphosphorylated PTEN and the PTEN-associated complex. Mol Cell Biol 2009; 29: 5377–5388.
Maccario H, Perera NM, Gray A, Downes CP, Leslie NR . Ubiquitination of PTEN. (phosphatase and tensin homolog) inhibits phosphatase activity and is enhanced by membrane targeting and hyperosmotic stress. J Biol Chem 2010; 285: 12620–12628.
Yilmaz ÖH, Morrison SJ . The PI-3kinase pathway in hematopoietic stem cells and leukaemia-initiating cells: A mechanistic difference between normal and cancer stem cells. Blood Cells Mol Dis 2008; 41: 73–76.
Falini B, Martelli MP, Bolli N, Ballanti S, Pettirossi V, Martelli MP . Acute myeloid leukemia with mutated nucleophosmin (NPM1): is it a distinct entity? Blood 2011; 117: 1109–1120.
Ferrara F, Izzo T, Criscuolo C, Riccardi C, Muccioli G, Viola A et al. Favorable outcome in patients with acute myelogenous leukemia with the nucleophosmin gene mutation autografted after conditioning with high-dose continuous infusion of idarubicin and busulfan. Biol Blood Marrow Transplant 2010; 16: 1018–1024.
Pasqualucci L, Liso A, Martelli MP, Bolli N, Pacini R, Tabarrini A et al. Mutated nucleophosmin detects clonal multilineage involvement in acute myeloid leukaemia: Impact on WHO classification. 2006 Blood 108: 4146–4155.
Leslie NR, Foti M . Non-genomic loss of PTEN function in cancer: not in my genes. Trends Pharmacol Sci 2011; 32: 131–140.
Acknowledgements
We are grateful to AM Gruszka (I.F.O.M. Milan) for kindly donating the T26 monoclonal antibody. This work was supported by AIRC (Associazione Italiana per la Ricerca sul Cancro), AIL (Associazione Italiana contro le Leucemie) and ANPCyT–FONCyT (Agencia Nacional de Promoción Científica y Tecnológica–Fondo para la Investigación Científica y Tecnológica, Argentina, PICT 2006-01865). This work was also supported by National Institutes of Health grants to PPP; KLC is a research fellow of the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET); NIN and FG are established investigators at CONICET.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Author Contributions
NIN designed, carried out and analyzed the experiments and co-wrote the manuscript. MSS carried out and analyzed the experiments and contributed in manuscript writing; MD and TO contributed with immunofluorescence analysis of patients samples; KLC, FG and IF carried out the experiments of western blot; FF performed the FRET analysis, analyzed the data and co-wrote the manuscript; GC contributed data analysis, interpretation of results and manuscript writing; LB, EC, and SA critically reviewed the manuscript, and amended the final report; FL-C. and PPP designed the study, supervised the research, co-wrote and edited the manuscript.
Rights and permissions
About this article
Cite this article
Noguera, N., Song, M., Divona, M. et al. Nucleophosmin/B26 regulates PTEN through interaction with HAUSP in acute myeloid leukemia. Leukemia 27, 1037–1043 (2013). https://doi.org/10.1038/leu.2012.314
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2012.314
Keywords
This article is cited by
-
MCL1 regulates AML cells metabolism via direct interaction with HK2. Metabolic signature at onset predicts overall survival in AMLs’ patients
Leukemia (2023)
-
The equilibrium of tumor suppression: DUBs as active regulators of PTEN
Experimental & Molecular Medicine (2022)
-
The deubiquitinase USP7 promotes HNSCC progression via deubiquitinating and stabilizing TAZ
Cell Death & Disease (2022)
-
Deubiquitinases in hematological malignancies
Biomarker Research (2021)
-
Opposing effects of NPM1wt and NPM1c mutants on AKT signaling in AML
Leukemia (2020)