Abstract
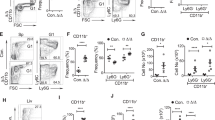
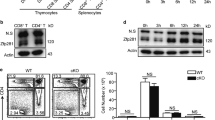
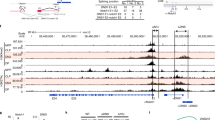
The Ets transcription factor PU.1, encoded by the gene Sfpi1, functions in a concentration-dependent manner to promote myeloid and B-cell development and has been implicated in myeloid and lymphoid leukemias. To determine the consequences of reducing PU.1 concentration during hematopoiesis, we analyzed mice with two distinct hypomorphic alleles of Sfpi1 that produce PU.1 at ∼20% (BN) or ∼2% (Blac) of wild-type levels. Myeloid development was impaired in these mice, but less severely than in Sfpi1 null mice. To identify the downstream target genes that respond to changes in PU.1 concentration, we analyzed ex vivo interleukin-3 dependent myeloid cell lines established from Sfpi1BN/BN, Sfpi1Blac/Blac and Sfpi1−/− fetal liver cells. Unexpectedly, many T-cell and natural killer cell genes were expressed in Sfpi1−/− cells and repressed in a dose-dependent manner in Sfpi1Blac/Blac and Sfpi1BN/BN cells. This pattern of dose-dependent T/NK-cell gene repression also occurred in ex vivo interleukin-7 dependent progenitor B cell lines. These results suggest that PU.1 functions in a concentration-dependent manner to repress T-cell and natural killer cell fates while promoting myeloid and B-cell fates.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Klemsz MJ, McKercher SR, Celada A, Van Beveren C, Maki RA . The macrophage and B cell-specific transcription factor PU.1 is related to the ets oncogene. Cell 1990; 61: 113–124.
McKercher SR, Torbett BE, Anderson KL, Henkel GW, Vestal DJ, Baribault H et al. Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO J 1996; 15: 5647–5658.
Scott EW, Simon MC, Anastasi J, Singh H . Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science 1994; 265: 1573–1577.
Spain LM, Guerriero A, Kunjibettu S, Scott EW . T-cell development in PU.1-deficient mice. J Immunol 1999; 163: 2681–2687.
Colucci F, Samson SI, DeKoter RP, Lantz O, Singh H, Di Santo JP . Differential requirement for the transcription factor PU.1 in the generation of natural killer cells versus B and T-cells. Blood 2001; 97: 2625–2632.
Mueller BU, Pabst T, Osato M, Asou N, Johansen LM, Minden MD et al. Heterozygous PU.1 mutations are associated with acute myeloid leukemia. Blood 2002; 100: 998–1007.
Vangala RK, Heiss-Neumann MS, Rangatia JS, Singh SM, Schoch C, Tenen DG et al. The myeloid master regulator transcription factor PU.1 is inactivated by AML1-ETO in t(8;21) myeloid leukemia. Blood 2003; 101: 270–277.
Rosenbauer F, Wagner K, Kutok JL, Iwasaki H, Le Beau MM, Okuno Y et al. Acute myeloid leukemia induced by graded reduction of a lineage-specific transcription factor, PU.1. Nat Genet 2004; 36: 624–630.
Cook WD, McCaw BJ, Herring C, John DL, Foote SJ, Nutt SL et al. PU.1 is a suppressor of myeloid leukemia, inactivated in mice by gene deletion and mutation of its DNA binding domain. Blood 2004; 104: 3437–3444.
Walter MJ, Park JS, Ries RE, Lau SK, McLellan M, Jaeger S et al. Reduced PU.1 expression causes myeloid progenitor expansion and increased leukemia penetrance in mice expressing PML-RARalpha. Proc Natl Acad Sci USA 2005; 102: 12513–12518.
Metcalf D, Dakic A, Mifsud S, Di Rago L, Wu L, Nutt S . Inactivation of PU.1 in adult mice leads to the development of myeloid leukemia. Proc Natl Acad Sci USA 2006; 103: 1486–1491.
Rosenbauer F, Owens BM, Yu L, Tumang JR, Steidl U, Kutok JL et al. Lymphoid cell growth and transformation are suppressed by a key regulatory element of the gene encoding PU.1. Nat Genet 2006; 38: 27–37.
Ross IL, Dunn TL, Yue X, Roy S, Barnett CJ, Hume DA . Comparison of the expression and function of the transcription factor PU.1 (Spi-1 proto-oncogene) between murine macrophages and B lymphocytes. Oncogene 1994; 9: 121–132.
DeKoter RP, Walsh JC, Singh H . PU.1 regulates both cytokine-dependent proliferation and differentiation of granulocyte/macrophage progenitors. EMBO J 1998; 17: 4456–4468.
Houston IB, Kamath MB, Schweitzer BL, Chlon TM, Dekoter RP . Reduction in PU.1 activity results in a block to B-cell development, abnormal myeloid proliferation, and neonatal lethality. Exp Hematol 2007; 35: 1056–1068.
DeKoter RP, Singh H . Regulation of B lymphocyte and macrophage development by graded expression of PU.1. Science 2000; 288: 1439–1441.
Dahl R, Walsh JC, Lancki D, Laslo P, Iyer SR, Singh H et al. Regulation of macrophage and neutrophil cell fates by the PU.1:C/EBPalpha ratio and granulocyte colony-stimulating factor. Nat Immunol 2003; 4: 1029–1036.
Laslo P, Spooner CJ, Warmflash A, Lancki DW, Lee HJ, Sciammas R et al. Multilineage transcriptional priming and determination of alternate hematopoietic cell fates. Cell 2006; 126: 755–766.
Back J, Allman D, Chan S, Kastner P . Visualizing PU.1 activity during hematopoiesis. Exp Hematol 2005; 33: 395–402.
Nutt SL, Metcalf D, D’Amico A, Polli M, Wu L . Dynamic regulation of PU.1 expression in multipotent hematopoietic progenitors. J Exp Med 2005; 201: 221–231.
Schuetze S, Stenberg PE, Kabat D . The Ets-related transcription factor PU.1 immortalizes erythroblasts. Mol Cell Biol 1993; 13: 5670–5678.
Anderson MK, Weiss AH, Hernandez-Hoyos G, Dionne CJ, Rothenberg EV . Constitutive expression of PU.1 in fetal hematopoietic progenitors blocks T-cell development at the pro-T-cell stage. Immunity 2002; 16: 285–296.
Rekhtman N, Radparvar F, Evans T, Skoultchi AI . Direct interaction of hematopoietic transcription factors PU.1 and GATA-1: functional antagonism in erythroid cells. Genes Dev 1999; 13: 1398–1411.
Rhodes J, Hagen A, Hsu K, Deng M, Liu TX, Look AT et al. Interplay of pu.1 and gata1 determines myelo-erythroid progenitor cell fate in zebrafish. Dev Cell 2005; 8: 97–108.
Galloway JL, Wingert RA, Thisse C, Thisse B, Zon LI . Loss of gata1 but not gata2 converts erythropoiesis to myelopoiesis in zebrafish embryos. Dev Cell 2005; 8: 109–116.
Stopka T, Amanatullah DF, Papetti M, Skoultchi AI . PU.1 inhibits the erythroid program by binding to GATA-1 on DNA and creating a repressive chromatin structure. EMBO J 2005; 24: 3712–3723.
Houston IB, Huang KJ, Jennings SR, DeKoter RP . PU.1 immortalizes hematopoietic progenitors in a GM-CSF-dependent manner. Exp Hematol 2007; 35: 374–384.
Schweitzer BL, DeKoter RP . Analysis of gene expression and Ig transcription in PU.1/Spi-B-deficient progenitor B cell lines. J Immunol 2004; 172: 144–154.
Pfaffl MW . A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 2001; 29: e45.
Markey MP, Bergseid J, Bosco EE, Stengel K, Xu H, Mayhew CN et al. Loss of the retinoblastoma tumor suppressor: differential action on transcriptional programs related to cell cycle control and immune function. Oncogene 2007; 26: 6307–6318.
Huang DW, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J et al. The DAVID gene functional classification tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol 2007; 8: R183.
Kodandapani R, Pio F, Ni CZ, Piccialli G, Klemsz M, McKercher S et al. A new pattern for helix-turn-helix recognition revealed by the PU.1 ETS-domain-DNA complex. Nature 1996; 380: 456–460.
Wingender E, Dietze P, Karas H, Knuppel R . TRANSFAC: a database on transcription factors and their DNA binding sites. Nucleic Acids Res 1996; 24: 238–241.
Hinrichs AS, Karolchik D, Baertsch R, Barber GP, Bejerano G, Clawson H et al. The UCSC Genome Browser Database: update 2006. Nucleic Acids Res 2006; 34 (Database issue): D590–D598.
Schwartz S, Elnitski L, Li M, Weirauch M, Riemer C, Smit A et al. MultiPipMaker and supporting tools: alignments and analysis of multiple genomic DNA sequences. Nucleic Acids Res 2003; 31: 3518–3524.
Walsh JC, DeKoter RP, Lee HJ, Smith ED, Lancki DW, Gurish MF et al. Cooperative and antagonistic interplay between PU.1 and GATA-2 in the specification of myeloid cell fates. Immunity 2002; 17: 665–676.
Takemoto CM, Yoon YJ, Fisher DE . The identification and functional characterization of a novel mast cell isoform of the microphthalmia-associated transcription factor. J Biol Chem 2002; 277: 30244–30252.
Nimmerjahn F, Ravetch JV . Fcgamma receptors: old friends and new family members. Immunity 2006; 24: 19–28.
Hu R, Sharma SM, Bronisz A, Srinivasan R, Sankar U, Ostrowski MC . Eos, MITF, and PU.1 recruit corepressors to osteoclast-specific genes in committed myeloid progenitors. Mol Cell Biol 2007; 27: 4018–4027.
Halbleib JM, Nelson WJ . Cadherins in development: cell adhesion, sorting, and tissue morphogenesis. Genes Dev 2006; 20: 3199–3214.
Rehli M, Lichanska A, Cassady AI, Ostrowski MC, Hume DA . TFEC is a macrophage-restricted member of the microphthalmia-TFE subfamily of basic helix-loop-helix leucine zipper transcription factors. J Immunol 1999; 162: 1559–1565.
Akagawa E, Muto A, Arai K, Watanabe S . Analysis of the 5′ promoters for human IL-3 and GM-CSF receptor alpha genes. Biochem Biophys Res Commun 2003; 300: 600–608.
Polli M, Dakic A, Light A, Wu L, Tarlinton DM, Nutt SL . The development of functional B lymphocytes in conditional PU.1 knock-out mice. Blood 2005; 106: 2083–2090.
Ye M, Ermakova O, Graf T . PU.1 is not strictly required for B cell development and its absence induces a B-2 to B-1 cell switch. J Exp Med 2005; 202: 1411–1422.
Henkel GW, McKercher SR, Maki RA . Identification of three genes up-regulated in PU.1 rescued monocytic precursor cells. Int Immunol 2002; 14: 723–732.
Cattoretti G, Shaknovich R, Smith PM, Jack H-M, Murty VV, Alobeid B . Stages of germinal center transit are defined by B cell transcription factor coexpression and relative abundance. J Immunol 2006; 177: 6930–6939.
Nutt SL, Heavey B, Rolink AG, Busslinger M . Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature 1999; 401: 556–562.
Pongubala JM, Northrup DL, Lancki DW, Medina KL, Treiber T, Bertolino E et al. Transcription factor EBF restricts alternative lineage options and promotes B cell fate commitment independently of Pax5. Nat Immunol 2008; 9: 203–215.
Tydell CC, David-Fung E-S, Moore JE, Rowen L, Taghon T, Rothenberg EV . Molecular dissection of prethymic progenitor entry into the T lymphocyte developmental pathway. J Immunol 2007; 179: 421–438.
Laiosa CV, Stadfeld M, Xie H, de Andres-Aguayo L, Graf T . Reprogramming of committed T-cell progenitors to macrophages and dendritic cells by C/EBPa and PU.1 transcription factors. Immunity 2006; 25: 731–744.
Acknowledgements
We thank the Affymetrix GeneChip Microarray Core (Cincinnati Children's Hospital Medical Center) for processing samples, Phil Sanford of the Gene Targeted Mouse Service (University of Cincinnati) for advice and H Leighton Grimes (Cincinnati Children's Hospital Medical Center) and Brock Schweitzer (University of Cincinnati) for helpful discussions. MBK is a PhD candidate at University of Cincinnati, and this work is submitted in partial fulfillment of the degree requirements. This work was supported by National Institutes of Health grant AI052175 and Ohio Cancer Research Associates grant 5407.
Contribution: MBK, IBH and RPD designed and performed experiments and collected data. AJJ and XZ performed experiments. MBK, SG and AGJ analyzed and interpreted data and performed statistical analyses. MBK and RPD drafted the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Conflict of interest/disclosure
The authors declare no competing financial interests.
Supplementary Information accompanies the paper on the Leukemia website (http://www.nature.com/leu)
Rights and permissions
About this article
Cite this article
Kamath, M., Houston, I., Janovski, A. et al. Dose-dependent repression of T-cell and natural killer cell genes by PU.1 enforces myeloid and B-cell identity. Leukemia 22, 1214–1225 (2008). https://doi.org/10.1038/leu.2008.67
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2008.67
Keywords
This article is cited by
-
Modest changes in Spi1 dosage reveal the potential for altered microglial function as seen in Alzheimer’s disease
Scientific Reports (2021)
-
Environmental sensing by mature B cells is controlled by the transcription factors PU.1 and SpiB
Nature Communications (2017)
-
The Transcription Factor PU.1 is a Critical Regulator of Cellular Communication in the Immune System
Archivum Immunologiae et Therapiae Experimentalis (2011)
-
Regulation of lymphoid versus myeloid fate 'choice' by the transcription factor Mef2c
Nature Immunology (2009)
-
Models of haematopoiesis: seeing the wood for the trees
Nature Reviews Immunology (2009)