Abstract
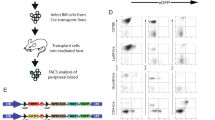
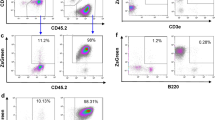
Insertional activation of cellular proto-oncogenes by replication-defective retroviral vectors can trigger clonal dominance and leukemogenesis in animal models and clinical trials. Here, we addressed the leukemogenic potential of vectors expressing interleukin-2 receptor common γ-chain (IL2RG), the coding sequence required for correction of X-linked severe combined immunodeficiency. Similar to conventional γ-retroviral vectors, self-inactivating (SIN) vectors with strong internal enhancers also triggered profound clonal imbalance, yet with a characteristic insertion preference for a window located downstream of the transcriptional start site. Controls including lentivirally transduced cells revealed that ectopic IL2RG expression was not sufficient to trigger leukemia. After serial bone marrow transplantation involving 106 C57Bl6/J mice monitored for up to 18 months, we observed leukemic progression of six distinct clones harboring γ-retroviral long terminal repeat (LTR) or SIN vector insertions in Evi1 or Prdm16, two functionally related genes. Three leukemic clones had single vector integrations, and identical clones manifested with a remarkably similar latency and phenotype in independent recipients. We conclude that upregulation of Evi1 or Prdm16 was sufficient to initiate a leukemogenic cascade with consistent intrinsic dynamics. Our study also shows that insertional mutagenesis is required for leukemia induction by IL2RG vectors, a risk to be addressed by improved vector design.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Cavazzana-Calvo M, Fischer A . Efficacy of gene therapy for SCID is being confirmed. Lancet 2004; 364: 2155–2156.
Gaspar HB, Parsley KL, Howe S, King D, Gilmour KC, Sinclair J et al. Gene therapy of X-linked severe combined immunodeficiency by use of a pseudotyped gammaretroviral vector. Lancet 2004; 364: 2181–2187.
Gaspar HB, Bjorkegren E, Parsley K, Gilmour KC, King D, Sinclair J et al. Successful reconstitution of immunity in ADA-SCID by stem cell gene therapy following cessation of PEG-ADA and use of mild preconditioning. Mol Ther 2006; 14: 505–513.
Ott MG, Schmidt M, Schwarzwaelder K, Stein S, Siler U, Koehl U et al. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat Med 2006; 12: 401–409.
Aiuti A, Slavin S, Aker M, Ficara F, Deola S, Mortellaro A et al. Correction of ADA-SCID by stem cell gene therapy combined with nonmyeloablative conditioning. Science 2002; 296: 2410–2413.
Li Z, Dullmann J, Schiedlmeier B, Schmidt M, von Kalle C, Meyer J et al. Murine leukemia induced by retroviral gene marking. Science 2002; 296: 497.
Seggewiss R, Pittaluga S, Adler RL, Guenaga FJ, Ferguson C, Pilz IH et al. Acute myeloid leukemia is associated with retroviral gene transfer to hematopoietic progenitor cells in a rhesus macaque. Blood 2006; 107: 3865–3867.
Modlich U, Kustikova OS, Schmidt M, Rudolph C, Meyer J, Li Z et al. Leukemias following retroviral transfer of multidrug resistance 1 (MDR1) are driven by combinatorial insertional mutagenesis. Blood 2005; 105: 4235–4246.
Cavazzana-Calvo M, Fischer A . Gene therapy for severe combined immunodeficiency: are we there yet? J Clin Invest 2007; 117: 1456–1465.
Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science 2003; 302: 415–419.
Board of the European Society of Gene and Cell Therapy. Case of leukaemia associated with X-linked severe combined immunodeficiency gene therapy trial in london. Hum Gene Ther 2008; 19: 3–4.
Kustikova O, Fehse B, Modlich U, Yang M, Dullmann J, Kamino K et al. Clonal dominance of hematopoietic stem cells triggered by retroviral gene marking. Science 2005; 308: 1171–1174.
Kustikova OS, Geiger H, Li Z, Brugman MH, Chambers SM, Shaw CA et al. Retroviral vector insertion sites associated with dominant hematopoietic clones mark ‘stemness’ pathways. Blood 2007; 109: 1897–1907.
Deichmann A, Hacein-Bey-Abina S, Schmidt M, Garrigue A, Brugman MH, Hu J et al. Vector integration is nonrandom and clustered and influences the fate of lymphopoiesis in SCID-X1 gene therapy. J Clin Invest 2007; 117: 2225–2232.
Schwarzwaelder K, Howe SJ, Schmidt M, Brugman MH, Deichmann A, Glimm H et al. Gammaretrovirus-mediated correction of SCID-X1 is associated with skewed vector integration site distribution in vivo. J Clin Invest 2007; 117: 2241–2249.
Aiuti A, Cassani B, Andolfi G, Mirolo M, Biasco L, Recchia A et al. Multilineage hematopoietic reconstitution without clonal selection in ADA-SCID patients treated with stem cell gene therapy. J Clin Invest 2007; 117: 2233–2240.
Modlich U, Bohne J, Schmidt M, von Kalle C, Knoss S, Schambach A et al. Cell-culture assays reveal the importance of retroviral vector design for insertional genotoxicity. Blood 2006; 108: 2545–2553.
Zychlinski D, Schambach A, Modlich U, Maetzig T, Meyer J, Grassman E et al. Physiological promoters reduce the genotoxic risk of integrating gene vectors. Mol Ther 2008; 16: 718–725.
Montini E, Cesana D, Schmidt M, Sanvito F, Ponzoni M, Bartholomae C et al. Hematopoietic stem cell gene transfer in a tumor-prone mouse model uncovers low genotoxicity of lentiviral vector integration. Nat Biotechnol 2006; 24: 687–696.
Shou Y, Ma Z, Lu T, Sorrentino BP . Unique risk factors for insertional mutagenesis in a mouse model of XSCID gene therapy. Proc Natl Acad Sci USA 2006; 103: 11730–11735.
Woods NB, Bottero V, Schmidt M, von Kalle C, Verma IM . Gene therapy: therapeutic gene causing lymphoma. Nature 2006; 440: 1123.
Thrasher AJ, Gaspar HB, Baum C, Modlich U, Schambach A, Candotti F et al. Gene therapy: X-SCID transgene leukaemogenicity. Nature 2006; 443: E5–E6; discussion E6–7.
Pike-Overzet K, de Ridder D, Weerkamp F, Baert MR, Verstegen MM, Brugman MH et al. Ectopic retroviral expression of LMO2, but not IL2Rgamma, blocks human T-cell development from CD34+ cells: implications for leukemogenesis in gene therapy. Leukemia 2007; 21: 754–763.
Pike-Overzet K, de Ridder D, Weerkamp F, Baert MR, Verstegen MM, Brugman MH et al. Gene therapy: is IL2RG oncogenic in T-cell development? Nature 2006; 443: E5; discussion E6–7.
Hildinger M, Abel KL, Ostertag W, Baum C . Design of 5′ untranslated sequences in retroviral vectors developed for medical use. J Virol 1999; 73: 4083–4089.
Schambach A, Wodrich H, Hildinger M, Bohne J, Krausslich HG, Baum C . Context dependence of different modules for posttranscriptional enhancement of gene expression from retroviral vectors. Mol Ther 2000; 2: 435–445.
Cavazzana-Calvo M, Hacein-Bey S, de Saint Basile G, Gross F, Yvon E, Nusbaum P et al. Gene therapy of human severe combined immunodeficiency (SCID)-X1 disease. Science 2000; 288: 669–672.
Schambach A, Mueller D, Galla M, Verstegen MM, Wagemaker G, Loew R et al. Overcoming promoter competition in packaging cells improves production of self-inactivating retroviral vectors. Gene Therapy 2006; 13: 1524–1533.
Dull T, Zufferey R, Kelly M, Mandel RJ, Nguyen M, Trono D et al. A third-generation lentivirus vector with a conditional packaging system. J Virol 1998; 72: 8463–8471.
Schmidt M, Hoffmann G, Wissler M, Lemke N, Mussig A, Glimm H et al. Detection and direct genomic sequencing of multiple rare unknown flanking DNA in highly complex samples. Hum Gene Ther 2001; 12: 743–749.
Relander T, Brun A, Hawley RG, Karlsson S, Richter J . Retroviral transduction of human CD34+ cells on fibronectin fragment CH-296 is inhibited by high concentrations of vector containing medium. J Gene Med 2001; 3: 207–218.
Huntly BJ, Gilliland DG . Cancer biology: summing up cancer stem cells. Nature 2005; 435: 1169–1170.
Pike-Overzet K, van der Burg M, Wagemaker G, van Dongen JJ, Staal FJ . New insights and unresolved issues regarding insertional mutagenesis in X-linked SCID gene therapy. Mol Ther 2007; 15: 1910–1916.
Akagi K, Suzuki T, Stephens RM, Jenkins NA, Copeland NG . RTCGD: retroviral tagged cancer gene database. Nucleic Acids Res 2004; 32 (Database issue): D523–D527.
Cattoglio C, Facchini G, Sartori D, Antonelli A, Miccio A, Cassani B et al. Hot spots of retroviral integration in human CD34+ hematopoietic cells. Blood 2007; 110: 1770–1778.
Wu X, Li Y, Crise B, Burgess SM . Transcription start regions in the human genome are favored targets for MLV integration. Science 2003; 300: 1749–1751.
Nucifora G, Laricchia-Robbio L, Senyuk V . EVI1 and hematopoietic disorders: history and perspectives. Gene 2006; 368: 1–11.
Barjesteh van Waalwijk van Doorn-Khosrovani S, Erpelinck C, van Putten WL, Valk PJ, van der Poel-van de Luytgaarde S, Hack R et al. High EVI1 expression predicts poor survival in acute myeloid leukemia: a study of 319 de novo AML patients. Blood 2003; 101: 837–845.
Buonamici S, Li D, Chi Y, Zhao R, Wang X, Brace L et al. EVI1 induces myelodysplastic syndrome in mice. J Clin Invest 2004; 114: 713–719.
Jin G, Yamazaki Y, Takuwa M, Takahara T, Kaneko K, Kuwata T et al. Trib1 and Evi1 cooperate with Hoxa and Meis1 in myeloid leukemogenesis. Blood 2007; 109: 3998–4005.
Cuenco GM, Ren R . Both AML1 and EVI1 oncogenic components are required for the cooperation of AML1/MDS1/EVI1 with BCR/ABL in the induction of acute myelogenous leukemia in mice. Oncogene 2004; 23: 569–579.
Du Y, Spence SE, Jenkins NA, Copeland NG . Cooperating cancer-gene identification through oncogenic-retrovirus-induced insertional mutagenesis. Blood 2005; 106: 2498–2505.
Louz D, van den Broek M, Verbakel S, Vankan Y, van Lom K, Joosten M et al. Erythroid defects and increased retrovirally-induced tumor formation in Evi1 transgenic mice. Leukemia 2000; 14: 1876–1884.
Mucenski ML, Taylor BA, Ihle JN, Hartley JW, Morse III HC, Jenkins NA et al. Identification of a common ecotropic viral integration site, Evi-1, in the DNA of AKXD murine myeloid tumors. Mol Cell Biol 1988; 8: 301–308.
Calmels B, Ferguson C, Laukkanen MO, Adler R, Faulhaber M, Kim HJ et al. Recurrent retroviral vector integration at the Mds1/Evi1 locus in nonhuman primate hematopoietic cells. Blood 2005; 106: 2530–2533.
Xinh PT, Tri NK, Nagao H, Nakazato H, Taketazu F, Fujisawa S et al. Breakpoints at 1p36.3 in three MDS/AML(M4) patients with t(1;3)(p36;q21) occur in the first intron and in the 5′ region of MEL1. Genes Chromosomes Cancer 2003; 36: 313–316.
Mochizuki N, Shimizu S, Nagasawa T, Tanaka H, Taniwaki M, Yokota J et al. A novel gene, MEL1, mapped to 1p36.3 is highly homologous to the MDS1/EVI1 gene and is transcriptionally activated in t(1;3)(p36;q21)-positive leukemia cells. Blood 2000; 96: 3209–3214.
Nishikata I, Sasaki H, Iga M, Tateno Y, Imayoshi S, Asou N et al. A novel EVI1 gene family, MEL1, lacking a PR domain (MEL1S) is expressed mainly in t(1;3)(p36;q21)-positive AML and blocks G-CSF-induced myeloid differentiation. Blood 2003; 102: 3323–3332.
Xiao Z, Zhang M, Liu X, Zhang Y, Yang L, Hao Y . MEL1S, not MEL1, is overexpressed in myelodysplastic syndromes patients with t(1;3)(p36;q21). Leuk Res 2006; 30: 332–334.
Hazourli S, Chagnon P, Sauvageau M, Fetni R, Busque L, Hebert J . Overexpression of PRDM16 in the presence and absence of the RUNX1/PRDM16 fusion gene in myeloid leukemias. Genes Chromosomes Cancer 2006; 45: 1072–1076.
Russell M, List A, Greenberg P, Woodward S, Glinsmann B, Parganas E et al. Expression of EVI1 in myelodysplastic syndromes and other hematologic malignancies without 3q26 translocations. Blood 1994; 84: 1243–1248.
Wieser R . The oncogene and developmental regulator EVI1: expression, biochemical properties, and biological functions. Gene 2007; 396: 346–357.
Morishita K . Leukemogenesis of the EVI1/MEL1 gene family. Int J Hematol 2007; 85: 279–286.
Evans-Galea MV, Wielgosz MM, Hanawa H, Srivastava DK, Nienhuis AW . Suppression of clonal dominance in cultured human lymphoid cells by addition of the cHS4 insulator to a lentiviral vector. Mol Ther 2007; 15: 801–809.
Acknowledgements
We thank Marina Cavazzana-Calvo and Alain Fischer (Hopital Necker, Paris) for the MFG.γc plasmid. This study was supported by grants from the Deutsche Forschungsgemeinschaft (DFG SPP1230 to ZL and CB), the European Union (CONSERT to CB), the Bundesministerium für Bildung und Forschung (BMBF TreatID to CB) and the National Cancer Institute (R01-CA107492-01A2, to CB). AS was supported by the Else-Kröner Foundation.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on the Leukemia website (http://www.nature.com/leu)
Supplementary information
Rights and permissions
About this article
Cite this article
Modlich, U., Schambach, A., Brugman, M. et al. Leukemia induction after a single retroviral vector insertion in Evi1 or Prdm16. Leukemia 22, 1519–1528 (2008). https://doi.org/10.1038/leu.2008.118
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2008.118
Keywords
This article is cited by
-
EVI1 expression in early-stage breast cancer patients treated with neoadjuvant chemotherapy
BMC Cancer (2022)
-
Evaluating the Safety of Retroviral Vectors Based on Insertional Oncogene Activation and Blocked Differentiation in Cultured Thymocytes
Molecular Therapy (2016)
-
Pigtailed macaques as a model to study long-term safety of lentivirus vector-mediated gene therapy for hemoglobinopathies
Molecular Therapy - Methods & Clinical Development (2014)
-
BET-independent MLV-based Vectors Target Away From Promoters and Regulatory Elements
Molecular Therapy - Nucleic Acids (2014)
-
Transduction of Fetal Mice With a Feline Lentiviral Vector Induces Liver Tumors Which Exhibit an E2F Activation Signature
Molecular Therapy (2014)