Abstract
PlexinA is a neuronal receptor protein that facilitates axon guidance during embryogenesis. This gene is associated with several neurological disorders including Alzheimer’s disease, Parkinson’s disease and autism. However, the effect of variants of PlexinA on brain structure remains unclear. We demonstrate that single-nucleotide polymorphisms within the intron and 3′-untranslated region segments of several human PlexinA genes alter the post-natal developmental trajectory of corpus callosum microstructure. This is the first demonstration that PLXNA mediation of neuroanatomical traits can be detected in humans using in vivo neuroimaging techniques. This result should encourage future research that targets specific disease-related polymorphisms and their relevant neural pathways.
Similar content being viewed by others
Introduction
PlexinA (PLXNA) is a family of cell-surface receptor proteins that guide developing axons to their targets within the central nervous system.1, 2 It is comprised of four genes, known as PlexinA1–A4.1, 3 Plexins form a receptor complex with neuropilins so as to bind semaphorins, thereby mediating the latter’s chemorepulsive influence on axonal growth-cone development.2, 4 Plexins are expressed throughout the developing nervous system.5 However, an important part of their role in brain development is to guide developing cortical axons across the midline, thereby contributing to the formation of the corpus callosum,6 a large white matter bundle that links the two cerebral hemispheres. Molecular signaling through this pathway maintains an orderly position of axons as they cross the midline at the corpus callosum, thereby allowing the axons to reach their appropriate homotopic targets in the contralateral hemisphere.7
Although most research on plexin has been conducted on Drosophila, C. elegans and rodents, recent studies on humans have linked common polymophisms in PLXNA4 with an increased risk of complex neurological disorders including Alzheimer’s disease,8 Parkinson’s disease9 and autism.10 The link between risk alleles and abnormal brain development in these diseases may be found in atypical expression levels or atypical structural isoforms of plexins. We present here the first study to examine the influence of variation in the PLXNA gene family on human white matter microstructure, with a focus on the developing corpus callosum.
Materials and methods
We used diffusion tensor imaging (DTI), a magnetic resonance-based technique that measures the diffusion of water,11 to assess white matter properties of the human corpus callosum during post natal development. A major advantage of DTI is that it permits noninvasive in vivo measurements of white matter microstructure. Axon membranes are only semipermeable to water, thereby creating a barrier to diffusion perpendicular to, but not parallel with, the main axis of white matter bundles. Fractional anisotropy (FA) is a DTI-derived measurement that quantifies the amount of diffusion parallel to, relative to perpendicular with, axon bundles.12 White matter bundles that take a straight path have high FA values, as the orientation of the resistance to diffusion is uniform throughout the bundle, whereas white matter bundles that take a tortuous path have lower FA values, as the direction of resistance to diffusion varies along the extent of the bundle.2
The data used in the present study were obtained from the Pediatric Imaging, Neurocognition and Genetics (PING) Study database (http://ping.chd.ucsd.edu). PING is a data resource comprised of highly standardized and carefully curated magnetic resonance imaging data, extensive genotyping data, and developmental and neuropsychological assessments for a large cohort of children three, collected across multiple sites. The PING database (version v0.4) was searched for single-nucleotide polymorphisms (SNPs) within the PLXNA1-4 genes. Genotype data for 1083 participants (aged 3–21 years, 521 female, 454 European, 110 African, 104 Asian, 64 South American, 12 Pacific Islands, 321 mixed ancestry, 18 other) were retrieved from the database along with FA for the corpus callosum for each participant. Data analysis was conducted in R v3.1 (Foundation for Statistical Computing, Austria, Vienna).13 Additive models14 were used to test whether FA varied in the corpus callosum as a function of genotype and/or the interaction of genotype with age, using age, sex, genetic ancestry and data-collection site as covariates.
Results
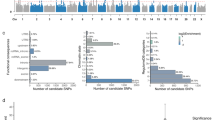
The PING database contained 13 SNPs across the four PLXNA genes (see Table 1). For all plexin genotypes, the corpus callosum FA increased throughout the course of development (Figure 1), demonstrating a trend consistent with previous research on callosal development.15 Three of 13 SNPs in our dataset exhibited a main effect of genotype, indicating differences in corpus callosum FA that persisted across development. Ten SNPs exhibited interactions between genotype and age, indicating variation in the developmental trajectory of corpus callosum FA. Only three of the 13 SNPs failed to predict FA.
Plexin-mediated developmental trajectory of the corpus callosum microstructure. (a–j) Plots of corpus callosum fractional anisotropy as a function of age for the 10 SNPs with significant main effects of genotype or interactions between genotype and age. Each plot includes the mean developmental trend (solid line) and one standard error above and below it (shaded area). The developmental trajectory for each genotype is plotted separately in each panel. In two cases (panels a and d), the developmental trajectory for homozygotes of the minor frequency allele was based on too few observations, resulting in unreliable estimates of the developmental trend for those alleles. Plots for SNPs that showed a significant main effect of genotype are marked with an asterisk. Lines showing a significant interaction between genotype and age are highlighted in bold. (k) Midsagittal magnetic resonance image of the human brain with the corpus callosum indicated by black arrows. A full color version of this figure is available at the Journal of Human Genetics journal online.
Discussion
Most of the PLXNA SNPs available in our database were introns and 3′-untranslated regions. Both types of regions regulate gene expression,16 suggesting that common polymorphisms within the PLXNA gene family may primarily affect the levels of gene expression. Introns can also regulate alternative splicing of exon sequences, resulting in different isoforms of a protein. Little is known about how plexin isoforms differ in function, and additional research is needed to elucidate their role in both typical and atypical brain development. However, SNPs can be statistically associated with nearby sequences through shared inheritance. This may explain the strong effects observed for a synonymous codon substitution for rs4679323. The SNPs that were analyzed in the present study may be statistically associated with nearby non-synonymous substitutions on exons that are not available in the PING database.
The present study demonstrates that natural variation in the PLXNA family has a measurable influence on the microstructure of the largest white matter tract in the human brain. This is particularly manifested as an abnormal developmental trajectory of fractional anisotropy, suggesting that plexin may have a prominent role in post natal regulation of white matter microstructure. The present study is an important step in understanding the relationship between these axon-guidance genes and complex neurological disorders in humans. Future research using similar in vivo methodologies should seek to link polymorphisms related to specific disorders with abnormal white matter microstructure in the brain regions related to those disorders. Complementary research using molecular genetic methodologies will be vital in elucidating the cellular mechanisms that drive the influence of PLXNA variation on white matter maturation in the human brain.
References
Tamagnone, L., Artigiani, S., Chen, H., He, Z., Ming, G. I. & Song, H. et al. Plexins are a large family of receptors for transmembrane, secreted, and GPI-anchored semaphorins in vertebrates. Cell 99, 71–80 (1999).
Takahashi, T., Fournier, A., Nakamura, F., Wang, L. H., Murakami, Y. & Kalb, R. G. et al. Plexin-neuropilin-1 complexes form functional semaphorin-3A receptors. Cell 99, 59–69 (1999).
Mauti, O., Sadhu, R., Gemayel, J., Gesemann, M. & Stoeckli, E. T. Expression patterns of plexins and neuropilins are consistent with cooperative and separate functions during neural development. BMC Dev. Biol. 17, 6–32 (2006).
Chisholm, A. & Tessier-Lavigne, M. Conservation and divergence of axon guidance mechanisms. Curr. Opin. Neurobiol. 9, 603–615 (1999).
Murakami, Y., Suto, F., Shimizu, M., Shinoda, T., Kameyama, T. & Fujisawa, H. Differential expression of Plexin-A subfamily members in the mouse nervous system. Dev. Dyn. 258, 246–258 (2001).
Gutekunst, C. -A., Stewart, E. N. & Gross, R. E. Immunohistochemical distribution of plexinA4 in the adult rat central nervous system. Front. Neuroanat. 4, 1–17 (2010).
Zhou, J., Wen, Y., She, L., Yui, Y.-N., Liu, L. & Richards, L. J. et al. Axon position within the corpus callosum determines contralateral cortical projection. Proc. Natl Acad. Sci. USA 110, e2714–e2723 (2013).
Jun, G., Asai, H., Drapeau, E., Park, J. H., Zeldich, E. & Chen, C. D. et al. Genetic variation in PLXNA4 associated with susceptibility of Alzheimer’s disease through tau phosphorylation. Alzheimer’s Dement. 9, 692–692 (2013).
Schulte, E. C., Stahl, I., Czamara, D., Ellwanger, D. C., Eck, S. & Graf, E. et al. Rare variants in PLXNA4 and Parkinson’s disease. PLoS ONE 8, e79145 (2013).
Suda, S., Iwata, K., Shimmura, C., Kameno, Y., Anitha, A. & Thanseem, I. et al. Decreased expression of axon-guidance receptors in the anterior cingulate cortex in autism. Mol. Autism 2, 14 (2011).
Basser, P. J. & Pierpaoli, C. Microstructural and physiological features of tissues elucidated by quantitative-diffusion-tensor. J. Magn. Reson. 213, 560–570 (1996).
Beaulieu, C. The basis of anisotropic water diffusion in the nervous system: a technical review. NMR Biomed. 15, 435–455 (2002).
R Core Team. R Foundation for Statistical Computing. R: A Language and Environment for Statistical Computing (2014). http://www.R-project.org/.
Wood, S. N. Generalized Additive Models: An Introduction with R, (CRC Press, Boca Raton, FL, USA, 2006).
Lebel, C., Gee, M., Camicioli, R., Wieler, M., Martin, W. & Beaulieu, C. Diffusion tensor imaging of white matter tract evolution over the lifespan. Neuroimage 60, 340–352 (2012).
Barrett, L. W., Fletcher, S. & Wilton, S. D. Regulation of eukaryotic gene expression by the untranslated gene regions and other non-coding elements. Cell Mol. Life Sci. 69, 3613–3634 (2012).
Acknowledgements
Data collection and sharing for this project was funded by the Pediatric Imaging, Neurocognition and Genetics Study (PING) via the National Institutes of Health Grant RC2DA029475 and the National Institute on Drug Abuse and the Eunice Kennedy Shriver National Institute of Child Health and Human Development. PING data are disseminated by the PING Coordinating Center at the Center for Human Development, University of California, San Diego. This work was supported by a grant to SB from the Natural Sciences and Engineering Research Council (NSERC) of Canada.
Author information
Authors and Affiliations
Consortia
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Belyk, M., Kraft, S., Brown, S. et al. PlexinA polymorphisms mediate the developmental trajectory of human corpus callosum microstructure. J Hum Genet 60, 147–150 (2015). https://doi.org/10.1038/jhg.2014.107
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2014.107
This article is cited by
-
An increase in Semaphorin 3A biases the axonal direction and induces an aberrant dendritic arborization in an in vitro model of human neural progenitor differentiation
Cell & Bioscience (2022)
-
Biallelic and monoallelic variants in PLXNA1 are implicated in a novel neurodevelopmental disorder with variable cerebral and eye anomalies
Genetics in Medicine (2021)
-
Proteome dynamics during postnatal mouse corpus callosum development
Scientific Reports (2017)
-
PLXNA3 Variant rs5945430 is Associated with Severe Clinical Course in Male Multiple Sclerosis Patients
NeuroMolecular Medicine (2017)