Abstract
Actinomycetes are a major source of bioactive secondary metabolites and are a focal point in the search for novel antimicrobial compounds that are needed to combat multidrug-resistant pathogens. Here, we report the discovery of several novel phenazine-type antibiotics produced by Kitasatospora sp. MBT66. These include the novel glycosylated endophenazines A–E (1–5), together with N-prenylated endophenazine F1 (6). Compounds 1 and 3 contain a 2′-O-methylation of the sugar moiety, which is rare in nature and reported for the first time in connection with phenazines. The structures of the new compounds were determined on the basis of their spectral data, including 1D and 2D NMR, HR-MS and the gene cluster responsible for the biosynthesis of phenazines was identified. All phenazine derivatives showed antimicrobial activity against the Gram-positive Bacillus subtilis, while compounds 1–3 and 5 also inhibited growth of the Gram-negative Escherichia coli.
Similar content being viewed by others
Introduction
Infections caused by multiple drug-resistant bacteria are a major threat to human health, both in the nosocomial environment and in the general community.1, 2, 3 It has become increasingly difficult to find novel antibiotics with efficacy against multidrug-resistant pathogens such as multidrug-resistant Mycobacterium tuberculosis, methicillin-resistant Staphylococcus aureus and multidrug-resistant Gram-negative pathogens.3 Indeed, despite intensive screening programs in the pharmaceutical industry, hardly any new drugs have been identified in this millennium, which underlines the urgent need for novel approaches to find antimicrobial drugs.4 Actinomycetes are Gram-positive mycelial bacteria with a complex multicellular life style.5, 6 They are industrially highly relevant because of their ability to produce a plethora of natural products, including anticancer, antifungal, anthelmantic and antibacterial drugs.7 Of all known naturally occurring antibiotics, some two-third is derived from actinomycetes, the majority of which are produced by streptomycetes.8 Sequencing of Streptomyces genomes established the presence of silent antibiotic biosynthetic gene clusters, suggesting that the potential of these organisms for novel drug production is much larger than originally anticipated.9 As an example, many novel gene clusters were identified even in the extensively studied model streptomycetes such as S. coelicolor,10 S. griseus11 and S. lividans.12 This has led to novel approaches to identifying specific conditions or agents that elicit the expression of antibiotics.13, 14, 15
In a recent study, we screened a collection of over 800 actinomycetes using 40 different growth conditions, which identified among others Kitasatospora sp. MBT66 that was shown to be a prolific producer of antibiotics with activity against Gram-positive and Gram-negative pathogens.16 Preliminary analysis predicted that it produced a number of compounds exhibiting similar chemical properties (TLC and HPLC-UV detection) to 1-carbomethoxy phenazine that was described in another actinomycete Streptomyces sp. MBT70.16 The phenazines are a group of heterocyclic, nitrogen-containing compounds isolated primarily from Pseudomonas and Streptomyces bacteria.17 They differ in chemical and physical properties according to the nature and/or position of functionalities, and thus give rise to a full spectrum of colors ranging from the deep red to the light blue.18 Phenazines are redox-active and can reduce molecular oxygen, leading to the generation of toxic reactive oxygen species and explaining their broad-spectrum antibiotic activity toward bacteria, fungi and plant and animal tissues.19, 20 Clofazimine is an example of a phenazine compound that is successfully applied in the clinic, where it is used for the treatment of leprosy, while clofazimine also has efficacy against the multidrug-resistant Mycobacterium tuberculosis.21 In addition, phenazines have multiple roles in the producing bacteria for survival and interaction with the environment, such as protection of plants by rhizobacteria against bacterial and fungal pathogens,22 reducing environmental Fe3+ to the more soluble Fe2+ to aid iron acquisition23 and in primary energy metabolism by mediating the reoxidation of NADH and utilization of pyruvate under oxygen-limiting conditions.24 Although more than 6000 phenazine-containing compounds have been described over the past century, fewer than 100 are of natural origin.25
Very few glycosylated phenazines have so far been found in nature, and these are without exception compounds with bioactivity that are derived from 6-deoxy-l-glycopyranosides.20 In this work, we report the isolation, structure determination and antimicrobial activity of novel glycosylated endophenazines (1–5) and an N-prenylated endophenazine (6) produced by Kitasatospora species MBT66, and present the gene cluster responsible for their biosynthesis.
Materials and methods
General experimental procedures
FT-IR was measured on Perkin-Elmer FT-IR Spectrometer Paragon 1000 (Perkin-Elmer, Waltman, MA, USA). UV measurements were performed using a Shimadzu UV mini-1240. NMR spectra were recorded in CH3OH-d4 on a Bruker DMX 500 MHz NMR and 600 MHz calibrated to a residual methanol-d4 at δ 3.30. HR-ESI-MS were collected on an Agilent 1200 series HPLC connected to a LTQ-Orbitrap spectrometer. Semi-preparative HPLC (pHPLC) was performed with a Shimadzu HPLC system and a 5-ml Rheodyne manual injection loop, equipped with a reversed-phase C18 coloumn (Phenomenex Luna C18 (2) 100 A 5 micron 250 × 10 mm, Phenomenex, Torrance, CA, USA). All the pHPLC experiments used 2 ml min−1 flow rate and fraction collection based on detected peak. TLC was performed with silica gel 60 (Merck, Darmstadt, Germany) plates using CHCl3/MeOH (10:1) and visualized with anisaldehyde/sulfuric acid reagent. All solvents and chemicals were of analytical and HPLC grade.
Microorganisms and culture conditions
Kitasatospora sp. MBT66 was isolated from Himalayan mountain soil as described previously.16 Cultivation was done as described previously.26 For solid-grown cultures, minimal media agar plates (MM) with glycerol and mannitol (1% w/v) as the carbon sources were used. Square agar plates (12 cm × 12 cm) were inoculated with 5 × 107 spores from a fresh spore suspension. In total 125 plates were used, and incubated at 30 °C for 6 days. Escherichia coli K12 and Bacillus subtilis 168 were used for screening of antimicrobial activity.
Extraction and isolation
Kitasatospora sp. MBT66 was fermented by stationary cultivation with a total volume of 5 l agar medium. After fermentation for 6 days, mycelia were combined and extracted with ethyl acetate by soaking in solvent overnight at room temperature. The extraction was evaporated under reduced pressure to obtain 2.0 g of a crude residue. This extract adsorbed by silica gel was first chromatographed on a Macroporous resin Diaion HP-20 from Supelco (Bellefonte, PA, USA) by eluting stepwise from H2O to MeOH. For monitoring, 20 μl of each fraction was subjected to TLC on premade silica gel plates (Merck) by using developing solvent system of chloroform and methanol (10:1). The 40% (v/v) methanol eluent showing interesting bands on TLC and moderate antibiotic activity in paper disc agar diffusion test with bacterium B. subtilis, was subsequently separated by silica gel (pore size 60 Å, 70−230 mesh, St Louis, MO, USA) column chromatography employing gradient elution from CHCl3 to MeOH, to give four subfractions (sfr.1–sfr.4). Sfr.2 was rechromatographed on Sephadex LH-20 (Sigma) coloumn, to purify the compound 2 (2.5 mg) and compound 6 (0.2 mg). Semi-preparative reversed-phase HPLC (Phenomenex Luna C18 (2) 100 Å 5 μm 250 × 10 mm) purification of sfr.1, eluting with a gradient of MeOH in H2O from 80–100% in 30 min, gave the pure compound 3 (tR=30.25 min, 0.3 mg) and 4 (tR=29.10 min, 0.2 mg). Separation of sfr.3 by HPLC using the gradient of MeOH in H2O from 50–80% in 30 min resulted in the purification of 5 (tR=21.25 min, 1.3 mg), and 7 (tR=28.50 min, 0.2 mg). Sfr.4 was similarly subjected to pHPLC employing the mobile phase MeOH in H2O from 50–80% in 30 min and led to the separation of 1 (tR=17.7 min, 0.35 mg).
Antimicrobial activity assays
Antimicrobial activity was determined basically following the methods described in reference.27 The respective compound (1–7) was dissolved in methonal (2 mg ml−1), and 25 μl of the solution was applied on a paper disk (d=6 mm). The disks were then placed onto an agar plate containing a soft agar overlay with either B. subtilis or E. coli. Ampicillin and streptomycin were used as positive controls, and the solvent methanol as the negative control. After incubation at 37 °C for 18 h, growth inhibition zones (in mm) were recorded as antimicrobial activity.
Results and discussion
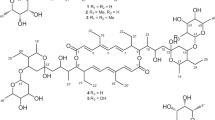
We previously performed a screen of a collection of some 800 actinomycetes obtained from mountain soils, so as to identify optimal laboratory conditions for antimicrobial secondary metabolites production.16 Kitasatospora sp. MBT6628 was one of our 96 most prolific antibiotic-producing identified actinomycetes,16 which was selected for further phytochemical investigation. Preliminary TLC detection of the crude extract of compounds produced by Kitasatospora sp. MBT66 on MM agar plates showed several yellow UV-active spots (254 nm) and orange fluorescence at 366 nm, which turned reddish-brown by staining with anisaldehyde/sulfuric acid reagent. Repeated chromatographic separation of upscaled extracts (2.0 g) coupled with subsequent spectroscopy data analysis resulted in the characterization of five previously undescribed rhamnosylated phenazines, endophenasides A–E (1–5) and one N-prenylated phenazine endophenazine F1 (6) with minor yield, together with the previously identified 1-carbomethoxy phenazine (7).16 The structures of the compounds are presented in Figure 1.
Resolution of the endophenaside structures
Compound 1 was obtained as a yellowish solid with a molecular formula of C20H21N3O6 established from high resolution mass [M + H]+ peak at m/z 400.15076. The 1H NMR (600 MHz, CH3OH-d4) of 1 (Table 1) showed six aromatic protons in two 1,2,3-trisubstituted aromatic spin systems, via δH 8.88 (dd, J=7.2, 1.2, Hz, H-2), 8.07 (dd, J=9.0, 7.2 Hz, H-3), and 8.54 (dd, J=9.0, 1.2 Hz, H-4), along with 7.69 (dd, J=7.2, 0.6 Hz, H-7), 7.97 (dd, J=9.0, 7.2 Hz, H-8), and 8.15 (dd, J=9.0, 0.6 Hz, H-9). Five protons in the range of δH 6.0–3.50, together with one methyl doublet at δH 1.26 (d, J=6.0 Hz, H3-6′) in high-magnetic field constituted another spin system by the confirmation of COSY experiment, which suggested the existence of a 6-deoxyhexapyranose sugar moiety (Figure 2). In addition, one distinct methoxyl δH 3.64 (s, H3-7′) was identified in the sugar region. The APT spectrum of compound 1 (Table 2) revealed 20 signals, which were assigned to carbonyl δC 168.7 (C-11), one oxygenated sp2 carbon δC 152.8 (C-6), six sp2 aromatic methine carbons δC 136.2 (C-2), 130.6 (C-3), 135.0 (C-4), 114.8 (C-7), 133.0 (C-8) and 123.5 (C-9) and five sp2 quaternary carbons (δC 130.1 (C-1), 143.1 (C-4a), 137.8 (C-5a), 143.7 (C-9a) and 141.7 (C-10a). Five oxygenated methine carbons δC 97.9 (C-1′), 81.5 (C-2′), 71.8 (C-3′), 73.7 (C-4′) and 70.9 (C-5′), together with one methyl group δC 17.7 (C-6′), were corresponding to the 6-deoxyhexapyranose by HSQC and APT spectra. The final structure of compound 1 was determined by analyses of the 2D NMR spectra (1H–1H COSY, HSQC and HMBC). The HMBC spectrum of compound 1 also suggested that it contained a 6-hydroxy-phenazine-1-carboxylic acid aglycone (H-2/C-11, C-10a, C-4; H-3/C-1; H-4/C-4a, C-2; H-7/C-9, C-6, C-5a; H-8/C-9a, C-6; and H-9/C-10a, C-7 and C-5a). The aglycone was also confirmed by comparison of the spectral data with the literature.29, 30, 31 Analysis of the COSY spectrum revealed one contiguous spin system formed by correlations of five oxygen-bearing methines and one doublet methyl signal at δH 1.39, confirming the deoxyhexapyranose. The connectivity of the 6-deoxysugar to the aglycone was confirmed from the HMBC long-range coupling of H-8 (δH 7.97) and H-1′ (δH 6.03) to the C-6 (δC 152.8) of the phenazine nucleus. The additional characteristic methoxy group was attached onto C-2′, based on two key HMBC correlation H-2′/C-7′ and H3-7′/C-2′ (Figure 2). The key HMBC correlation H-2/C-11 (δC 168.7) established a carbonyl substituent at C-1. To satisfy the molecular formula C20H21N3O6 deduced by HRMS, the substituent at C-1 had to be an amide instead of a carboxyl. Consequently, the structure of 1 was assigned as a new hydroxyphenzine-1-carboxyamide glycoside and designed as endophenaside A (Figure 1).
Compounds 2–4 and 6 possess the basic phenazine skeleton in comparison with 1, and the structure elucidation was done by NMR and HRMS (Supplementary Data File 1; for physical properties of the new compounds and all supporting spectra for the structure elucidation, see Supplementary Data Files 2–3). Notably, Heine et al.27 recently described endophenazines produced by Kitasatospora strain HK714, which were similar to but distinct from compound 1. Intensive spectroscopy data analysis for compounds 2–4 showed that their aglycone moieties were identical to those of previously described endophenazines,27 except that compounds 2–4 carried an additional sugar moiety. Compound 6 (endophenazine F1) resembled endophenazine F, but containd a hydroxylated prenyl group at the N-5 position, with the hydroxyl group at a terminal methyl group.
Compound 5 (endophenazine E) featured a phenazine nucleus and a 6-deoxyhexopyranose sugar as shown by 1H NMR. However, seven aromatic protons with overlapped and complicated coupling split patterns suggested a monosubstituted instead of a disubstituted pattern. The key HMBC correlation from sugar anomeric proton H-1′ (δH 6.39) to C-11 (166.7) confirmed the connectivity of a sugar substituent to the phenazine core through a carbonyl group. Furthermore, endophenaside E (5) was found to be unstable and prone to methanolysis to generate 1-carbomethoxy phenazine (7) during NMR experiments (Supplementary Figure S1).
The relative stereochemistry of the 6-deoxyhexopyranose in compounds 1–5 was determined on the basis of NOE observations and coupling constants.30, 32 The small coupling constant (J1′,2′ 1.5 Hz) between the anomeric proton and H-2′ suggested that H-1′ and H-2′ were both in equatorial positions and assigned the relative configuration of H-1′ to be α, since the NOE was not observed either between H-1′ and H-3′ or between H-1′ and H-5′. The NOE observed between H-3′ and H-5′ as well as the coupling constant between H-4′ and H-5′ (J4′,5′ 9.5 Hz) indicated that they were axially oriented. Based on further comparison with NMR data reported previously,30 6-deoxyhexopyranose in compounds 1–5 was identified as an α-rhamnose (Figure 3). As mentioned above, few glycosylated phenazines have so far been found in nature, most of which are rhamnosides with significant bioactivity.30, 31 Endophenasides A (1) and C (3) are the first example for 2′-O-methylation of the rhamnose moiety in the phenazine glycosides.
The HMBC spectrum does not allow distinguishing between 1,6- or 1,9-disubstituted phenazine.27, 30, 31, 32, 33 Indeed, the prenyl- and the carboxyl group on the phenazine core of compounds 1–4, could be either 1,6- or 1,9-disubstituted. However, the presence of a 1,9-disubstituted pattern is deduced from previous analysis of the aglycone moiety27 that is also present in compounds 2–4. Moreover, close scrutiny of Kitasatospora sp. MBT66 by UHPLC-Q-TOF analysis showed that Kitasatospora sp. MBT66 could in fact also produce most of the previously described aglycons (endophenazines) produced by Kitasatospora sp. HK714, albeit with a low yield (Supplementary Figure S2 and Supplementary Table S1). Conversely, the disubstitution pattern in compound 1 was tentatively proposed as [1,6-], which was deduced from the possible biosynthesis pathway (see below).
Antimicrobial activity of the endophenasides
Antimicrobial properties of compounds 1–7 were evaluated by agar diffusion assay, against the Gram-positive indicator strain B. subtilis 168 and the Gram-negative E. coli K12 (Table 3). Compound 5 showed highest antibiotic activity against both strains, while compounds 4 and 6 only inhibited B. subtilis and not E. coli. These antimicrobial activities are similar to the data published previously for the aglycones.27 Remarkably, the glycosylation enhanced the activity of compounds 2 and 3 against Gram-negative E. coli, in contrast to what was reported for the corresponding aglycone. Taken together, our data show that the novel endophenasides described here have significant antimicrobial activity, whereby the glycosylation did not inhibit the bioactivity.
The phenazine biosynthetic gene cluster
Annotation of the genome sequence of Kitasatospora sp. MBT66 (Genbank accession number JAIY0000000028) identified a gene cluster of around 25 kb that is likely responsible for the production of phenazine-type compounds. The gene products of the biosynthetic gene cluster (Table 4) were nearly identical to those of the endophenazine biosynthetic gene cluster of Kitasatospora sp. HKI 714 (epaA-epaU;27 GenBank accession number: KJ207079). It was proposed that EpaA catalyzes the prenylation at C-9 and/or N-5 positions of phenazine-1-carboxylic acid, similar to the function of enzyme PpzP in Streptomyces anulatus,34 which generated the [1,9-] substitution mode on the phenazine nucleus. From the perspective of biosynthesis, the formation of phenazine-1-carboxylic acid not only depends on the enzymes encoded by the gene cluster, but also involves spontaneous reaction without enzymatic catalysis.19, 35 The carboxyl group derived from chorismate is likely cleaved by molecular oxygen through oxidative decarboxylation, which would be alternatively replaced by a hydroxyl group in the aqueous environment (Figure 4). This may explain why 6-hydroxyphenazine-1-carboxylic acid derivatives were frequently identified,30, 31, 36, 37, 38 while 9-hydroxyphenazine-1-carboxylic acid39, 40 was less often found.
Proposed biogenesis of phenazine derivatives. Enzymes catalyzing the reaction correspond to Table 4. This biosynthesis pathway of phenazines was summarized according to references.19, 20, 35, 42 Reactions leading to the formation of the phenazine core are highlighted by a dashed box, which includes spontaneous reaction without enzymatic catalysis.19, 35 A yet uncharacterized glycosyltransferase and methyltransferase then give rise to the new glycosylated endophenazines (1–5) described in this study.
Besides the glycosylated endophenasides, we were able to show that Kitasatospora sp. MBT66 also produces small amounts of the same endophenazines as produced by Kitasatospora sp. HKI 714 (Supplementary Figure S2 and Supplementary Table S1). We expect that the two strains are closely related kitasatosporae, a genus closely related to Streptomyces in the family of Streptomycetaceae.41 This should be resolved by comparison of the genomes and phenotypic characteristics of the two strains. It is yet unclear which genes are responsible for the glycosylation of the endophenazine aglycones, as no such genes were found within or in close proximity of the epa gene cluster (Table 4). Therefore, the genes responsible for the transfer and methylation of the rhamnose moiety to either the carboxyl or the hydroxyl group of the phenazines should be located elsewhere on the chromosome. Research to identify the genes and enzymes responsible for the glycosylation of the endaphenazines is currently in progress.
Accession codes
References
Giske, C. G., Monnet, D. L., Cars, O. & Carmeli, Y. Clinical and economic impact of common multidrug-resistant Gram-negative bacilli. Antimicrob. Agents Chemother. 52, 813–821 (2008).
Spellberg, B. et al. The epidemic of antibiotic-resistant infections: a call to action for the medical community from the Infectious Diseases Society of America. Clin. Infect. Dis. 46, 155–164 (2008).
Rice, L. B. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: no ESKAPE. J. Infect. Dis. 197, 1079–1081 (2008).
Payne, D. J., Gwynn, M. N., Holmes, D. J. & Pompliano, D. L. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat. Rev. Drug Discov. 6, 29–40 (2007).
Claessen, D., Rozen, D. E., Kuipers, O. P., Søgaard-Andersen, L. & van Wezel, G. P. Bacterial solutions to multicellularity: a tale of biofilms, filaments and fruiting bodies. Nat. Rev. Microbiol. 12, 115–124 (2014).
Flärdh, K. & Buttner, M. J. Streptomyces morphogenetics: dissecting differentiation in a filamentous bacterium. Nat. Rev. Microbiol. 7, 36–49 (2009).
Hopwood, D. A. Streptomyces in Nature and Medicine: the Antibiotic Makers, (Oxford University Press, New York, (2007).
Miyadoh, S. Research on antibiotic screening in Japan over the last decade: a producing microorganism approach. Actinomycetol 7, 100–106 (1993).
Challis, G. L. & Hopwood, D. A. Synergy and contingency as driving forces for the evolution of multiple secondary metabolite production by Streptomyces species. Proc. Natl Acad. Sci. USA 100, 14555–14561 (2003).
Thomson, N. R. et al. Complete genome sequence of the model actinomycete Streptomyces. Nature 3, 141–147 (2002).
Ohnishi, Y. et al. Genome sequence of the streptomycin-producing microorganism Streptomyces griseus IFO 13350. J. Bacteriol. 190, 4050–4060 (2008).
Cruz-Morales, P. et al. The genome sequence of Streptomyces lividans 66 reveals a novel tRNA-dependent peptide biosynthetic system within a metal-related genomic island. Genome Biol. Evol. 5, 1165–1175 (2013).
Craney, A., Ozimok, C., Pimentel-Elardo, S. M., Capretta, A. & Nodwell, J. R. Chemical perturbation of secondary metabolism demonstrates important links to primary metabolism. Chem. Biol. 19, 1020–1027 (2012).
Van Wezel, G. P., McKenzie, N. L. & Nodwell, J. R. Applying the genetics of secondary metabolism in model actinomycetes to the discovery of new antibiotics. Methods Enzymol 458, 117–141 (2009).
Zhu, H., Sandiford, S. K. & van Wezel, G. P. Triggers and cues that activate antibiotic production by actinomycetes. J. Ind. Microbiol. Biotechnol. 41, 371–386 (2014).
Zhu, H. et al. Eliciting antibiotics active against the ESKAPE pathogens in a collection of actinomycetes isolated from mountain soils. Microbiology 160, 1714–1725 (2014).
Abken, H., Tietze, M. & Brodersen, J. Isolation and characterization of methanophenazine and function of phenazines in membrane-bound electron transport of Methanosarcina mazei Gö1. J. Bacteriol. 180, 2027–2032 (1998).
Price-Whelan, A., Dietrich, L. E. P. & Newman, D. K. Rethinking ‘secondary’ metabolism: physiological roles for phenazine antibiotics. Nat. Chem. Biol. 2, 71–78 (2006).
Ahuja, E. G. et al. PhzA/B catalyzes the formation of the tricycle in phenazine biosynthesis. J. Am. Chem. Soc. 130, 17053–17061 (2008).
Laursen, J. B. & Nielsen, J. Phenazine natural products: biosynthesis, synthetic analogues, and biological activity. Chem. Rev. 104, 1663–1686 (2004).
Cholo, M. C., Steel, H. C., Fourie, P. B., Germishuizen, W. A. & Anderson, R. Clofazimine: current status and future prospects. J. Antimicrob. Chemother. 67, 290–298 (2012).
Cook, R. & Thomashow, L. Molecular mechanisms of defense by rhizobacteria against root disease. Proc. Natl Acad. Sci. USA 92, 4197–4201 (1995).
Hernandez, ME, Kappler, A & Newman, DK Phenazines and other redox-active antibiotics promote microbial mineral reduction. Appl. Environ. Microbiol. 70, 921–928 (2004).
Wang, Y., Kern, S. E. & Newman, D. K. Endogenous phenazine antibiotics promote anaerobic survival of Pseudomonas aeruginosa via extracellular electron transfer. J. Bacteriol. 192, 365–369 (2010).
Mavrodi, D. V., Blankenfeldt, W. & Thomashow, L. S. Phenazine compounds in fluorescent Pseudomonas spp. biosynthesis and regulation. Annu. Rev. Phytopathol. 44, 417–445 (2006).
van Wezel, G. P. et al. A new piece of an old jigsaw: glucose kinase is activated posttranslationally in a glucose transport-dependent manner in Streptomyces coelicolor A3(2. J. Ind. Microbiol. Biotechnol. 12, 67–74 (2007).
Heine, D., Martin, K. & Hertweck, C. Genomics-Guided Discovery of Endophenazines from Kitasatospora sp. HKI 714. J. Nat. Prod. 77, 1083–1087 (2014).
Girard, G. et al. Analysis of novel kitasatosporae reveals significant evolutionary changes in conserved developmental genes between Kitasatospora and Streptomyces. Antonie Van Leeuwenhoek 106, 365–380 (2014).
Romer, A. 1H NMR spectra of substituted phenazines. Org. Magn. Reson 19, 66–68 (1982).
Abdelfattah, M. S., Toume, K. & Ishibashi, M. Isolation and structure elucidation of izuminosides A-C: a rare phenazine glycosides from Streptomyces sp. IFM 11260. J. Antibiot. (Tokyo) 64, 271–275 (2011).
Rusman, Y., Oppegard, L. M., Hiasa, H., Gelbmann, C. & Salomon, C. E. Solphenazines A-F, glycosylated phenazines from Streptomyces sp. strain DL-93. J. Nat. Prod. 76, 91–96 (2013).
Kato, S., Shindo, K. & Yamagishi, Y. Phenazoviridin, a novel free radical scavenger from Streptomyces sp. Taxonomy, fermentation, isolation, structure elucidation and biological properties. J. Antibiot. (Tokyo) 46, 1485–1493 (1993).
Krastel, P. & Zeeck, A. Endophenazines A-D, New Phenazine Antibiotics from the Athropod Associated Endosymbiont Streptomyces anulatus. II. Structure elucidation. J. Antibiot. (Tokyo) 55, 801–806 (2002).
Saleh, O., Gust, B., Boll, B., Fiedler, H.-P. & Heide, L. Aromatic prenylation in phenazine biosynthesis: dihydrophenazine-1-carboxylate dimethylallyltransferase from Streptomyces anulatus. J. Biol. Chem. 284, 14439–14447 (2009).
Rui, Z. et al. Insights into a divergent phenazine biosynthetic pathway governed by a plasmid-born esmeraldin gene cluster. Chem. Biol. 19, 1116–1125 (2012).
Ohlendorf, B., Schulz, D., Erhard, A., Nagel, K. & Imhoff, J. F. Geranylphenazinediol, an acetylcholinesterase inhibitor produced by a Streptomyces species. J. Nat. Prod. 75, 1400–1404 (2012).
Abdelfattah, M. S., Kazufumi, T. & Ishibashi, M. Izumiphenazines A-C: isolation and structure elucidation of phenazine derivatives from Streptomyces sp. IFM 11204. J. Nat. Prod. 73, 1999–2002 (2010).
Abdelfattah, M. S., Toume, K. & Ishibashi, M. Izumiphenazine D, a new phenazoquinoline N-oxide from Streptomyces sp. IFM 11204. Chem. Pharm. Bull. (Tokyo) 59, 508–510 (2011).
Fotso, S. et al. Modified phenazines from an Indonesian Streptomyces sp. J. Nat. Prod. 73, 472–575 (2010).
Gerber, N. New microbial phenazines. J. Heterocycl. Chem. 6, 4–7 (1969).
Girard, G. et al. A novel taxonomic marker that discriminates between morphologically complex actinomycetes. Open Biol 3, 130073 (2013).
Schneemann, I., Wiese, J., Kunz, A. L. & Imhoff, J. F. Genetic approach for the fast discovery of phenazine producing bacteria. Mar. Drugs 9, 772–789 (2011).
Acknowledgements
The work was supported by a PhD Fellowship from the China Scholarship Council to CSW, and by grants from the Netherlands Technology Foundation (10467 and 11992) to GPvW. Physics data of compounds 1–6, Structure elucidation of compounds 2–4 and 6, instability of compound 5 in methanol, detection of endophenazines, as well as HRESIMS, IR, UV, 1D and 2D NMR spectra of compounds 1–6 are available.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on The Journal of Antibiotics website
Supplementary information
Rights and permissions
About this article
Cite this article
Wu, C., van Wezel, G. & Hae Choi, Y. Identification of novel endophenaside antibiotics produced by Kitasatospora sp. MBT66. J Antibiot 68, 445–452 (2015). https://doi.org/10.1038/ja.2015.14
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2015.14
This article is cited by
-
Chemical Components of Selaginella trichoclada
Chemistry of Natural Compounds (2021)
-
Microbial diversity of a Himalayan forest and characterization of rare actinomycetes for antimicrobial compounds
3 Biotech (2019)
-
Genus Kitasatospora, taxonomic features and diversity of secondary metabolites
The Journal of Antibiotics (2017)
-
PhzA, the shunt switch of phenazine-1,6-dicarboxylic acid biosynthesis in Pseudomonas chlororaphis HT66
Applied Microbiology and Biotechnology (2017)
-
Metabolomics-guided analysis of isocoumarin production by Streptomyces species MBT76 and biotransformation of flavonoids and phenylpropanoids
Metabolomics (2016)