Abstract
A novel actinomycete, strain KC-061T was isolated from a soil sample collected from Angthong Islands National Park, Surat Thani province, Thailand. Strain KC-061T was identified as a member of the genus Nonomuraea through the use of a polyphasic approach. Phylogenetic analyses based on 16S rRNA gene sequencing, strain KC-061T belongs to the genus Nonomuraea and was closely related to ‘N. monospora’ PT708T (99.3%), N. rhizophila YIM 67092T (98.6%), N. dietziae DSM 44320T (98.5%) and N. rosea GW 12687T (98.3%). On the basis of phenotypic characteristics, DNA–DNA relatedness and phylogenetic distinctiveness, the novel isolate was identified as representing a novel species of the genus Nonomuraea, for which the name Nonomuraea thailandensis sp. nov. (type strain KC-061T =JCM 18408T=KCTC 29074T =PCU 327T) is proposed.
Similar content being viewed by others
Introduction
The genus Nonomuraea belongs to the family Streptosporangiaceae, which was first proposed by Zhang et al.1 Members of the genus Nonomuraea are aerobic, Gram-positive, non-acid-fast, non-motile actinomycetes that can form extensively branched substrate and aerial mycelia. At the time of writing, 30 Nonomuraea species names and 2 recognized subspecies have been validly published.2 In addition, some new species have been described in the past few years, namely N. antimicrobica,3 N. candida,4 N. endophytica,5 N. maritima,6 N. rhizophila,7 N. rosea,8 N. wenchangensis,9 N. jiangxiensis10 and N. soli.11 It has been reported that the strains that belong to the genus Nonomuraea produce various types of secondary metabolites; for example, glycopeptide antibiotic A40926,12, 13 myxochelin A,14 anthelmintic macrolactams, fluvirucin B0, Sch 38516/fluvirucin B1 and Sch 39185/fluvirucin B3,15 a novel cyclic tetrapeptide WSS222016 and new cyclic tetrapeptides.17 The strain KC-061T was isolated from the southern area of Thailand, which also produce madurahydroxylactone.18, 19 In this paper, we describe the characterization and classification of the strain in the genus Nonomuraea.
Results and discussion
Morphological, cultural, physiological and biochemical characteristics
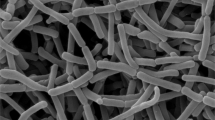
Strain KC-061T grew well on ISP 2, 3, 4, 5 and 7 media, YS agar and nutrient agar. The aerial mycelia appeared white on ISP 2 medium, whitish purple on ISP 3 medium and pale orange on ISP 5 and 7 media. No aerial mycelia were formed when the strain cultivated on ISP 4 and 6 media, YS agar and nutrient agar. The substrate mycelium branched extensively and the colors on various media were reddish brown to yellowish brown. Brown soluble pigments on ISP 2 and 3 media, YS agar and nutrient agar were produced (Table 1). The aerial hyphae produced long and spiraled spore chains, which had spiral bearing >10 spores (Figure 1). The strain grew at 14 to 38°C (optimally at 20 to 30°C) and at pH 6.0 to 11.0 (optimally pH at pH 7 to 9) and tolerated up to 4% NaCl. Nitrate was reduced to nitrite. Casein and starch were hydrolyzed but gelatin was not liquefied. Milk was peptonized weakly and was not coagulated. The strain KC-061T utilized D-glucose, myo-inositol, D-mannitol, melibiose, raffinose, L-rhamnose and D-xylose but did not utilize sucrose. Enzyme activities of the API ZYM system were positive for alkaline phosphatase, leucine arylamidase, α-chymotrypsin, acid phosphatase, naphthol-AS-BI-phosphohydrase, α-glucosidase, β -glucosidase and N-acetyl-β-glucosaminidase and weakly positive for esterase (C4), esterase lipase (C8), valine arylamidase and α-mannosidase but negative for cystine arylamidase, trypsin, α-galactosidase and β-galactosidase (Table 2).
Chemotaxonomy
Strain KC-061T contained meso-diaminopimelic acid isomer as diagnostic diamino acid and galactose, mannose, madurose and ribose as diagnostic sugars in whole cells. The N-acyl type of muramic acid was acetyl. Mycolic acids were not detected (data not shown). The predominant menaquinone was MK-9 (H4) (80%) and the minor menaquinones were MK-9 (H6) (10%), MK-9 (H2) (7%), MK-9 (H0) (2%) and MK-10 (H4) (1%). Strain KC-061T contained the same predominant menaquinone of MK-9 (H4) as strain ‘N. monospora’ PT708T;21 however, the minor amounts of MK-9 (H0) and MK-10 (H4) was absent in strain ‘N. monospora’ PT708T.21 Diphosphatidylglycerol, phosphatidylmonomethylethanolamine, phosphatidylethanolamine, hydroxy-phosphatidylmonomethylethanolamine, hydroxy-phosphatidylethanolamine, phosphatidylglycerol, ninhydrin positive phosphoglycolipid and unknown phospholipid (PL1) were detected as phospholipids. Strain KC-061T contained the same polar lipids as strain ‘N. monospora PT708T 21 but the unknown phospholipid (PL2) was not detected as found in ‘N. monospora’ PT708T (Figure 2). The major fatty acids were iso-C16:0 (25.3%), 10-methyl-C17:0 (13.4%), C17:1 ω8c (7.7%), C16:1 ω7c/C16:1 ω6c (7.0%), C16:0 (6.9%), iso-C16:1 G (6.4%), iso-C15:0 (4.8%), 10-methyl-C16:0 (4.0%) and C16:0 2-OH (3.8%). Strain KC-061T contained the same cellular fatty acid profiles as Nonomuraea species but it showed different in the amount of fatty acids (Table 3). The G+C content of genomic DNA was 72.4 mol%. Chemotaxonomic analyses confirmed that strain KC-061T exhibited typically chemical characteristics of members of the genus Nonomuraea.
Phospholipid compositions of strain KC-061T (a) and Nonomuraea monospora TISTR 1910T (b) (The TLC plates were sprayed with molybdatophosphoric acid). Diphosphatidylglycerol (DPG), phosphatidylmonomethylethanolamine (PME), phosphatidylethanolamine (PE), hydroxy-phosphatidylmonomethylethanolamine (OH-PME), hydroxy-phosphatidylethanolamine (OH-PE), phosphatidylglycerol (PG), ninhydrin positive phosphoglycolipid (NPG) and unknown phospholipids (PL1 and PL2). A full color version of this figure is available at The Journal of Antibiotics journal online.
Phylogenetic analysis
The 16S rRNA gene sequence analysis showed that strain KC-061T belongs to the genus Nonomuraea. The 16S rRNA gene sequence similarity values between the strain KC-061T and the type strains in the genus Nonomuraea ranged from 95.4 to 99.3%. The phylogenetic trees based on 16S rRNA gene sequences of strain KC-061T and the type strains of members in the genus Nonomuraea revealed that strain KC-061T constructed a cluster with ‘N. monospora’ PT708T, N. rhizophila YIM 67092T and N. rosea GW 12687T (Figure 3). The strain KC-061T showed high 16S rRNA gene sequence similarity values to ‘N. monospora’ PT708T 21 (99.3%), N. rhizophila DSM 45382T (98.6%), N. dietziae NBRC 14039T (98.5%) and N. rosea DSM 45177T (98.3%). These results indicate that strain KC-061T is the closest with ‘N. monospora’ PT708T. However, ‘N. monospora’ PT708T producing single spore is clearly distinguished from the strain KC-061T, which produced spiral spore chains.21
Neighbor-joining tree based on 16S rRNA gene sequences showing relationship between KC-061T and members of the genus Nonomuraea. Only bootstrap values above 50% (percentages of 1000 replications) are indicated. •, branch also recovered in the maximum-parsimony tree; *, branch also recovered in the maximum-likelihood tree; Bar, 0.01 nucleotide substitutions per site.
DNA–DNA hybridization
DNA–DNA relatedness values between strain KC-061T and strains ‘N. monospora’ PT708T, N. rhizophila DSM 45382T, N. dietziae NBRC 14309T and N. rosea DSM 45177T were 47±2.9%, 55±2.5%, 48±9.0% and 54±8.4%, respectively and reciprocally, strain ‘N. monospora’ PT708T showed 54±4.8%, 50±2%, 39±1.6% and 46±4.8% DNA–DNA relatedness to KC-061T, N. rhizophila DSM 45382T, N. dietziae NBRC 14309T and N. rosea DSM 45177T, respectively. These values are obtained from three independent determinations and are below the 70% cutoff point recommended by Wayne et al.22 for assigning strains to the same species and confirms the separation of strain KC-061T from its closely related phylogenetic neighbors. Therefore, strain KC-061T clearly represents a novel species of the genus Nonomuraea.
Conclusion
The results of phylogenetic and phenotypic characteristics of strain KC-061T are consistent with its classification within the genus Nonomuraea. Futhermore, there were many cultural, physiological and biochemical features that differed between strain KC-061T and the most closely related type strains, ‘N. monospora’ PT708T, N. rhizophila DSM 45382T, N. dietziae NBRC 14309T and N. rosea DSM 45177T, such as differences in production and color of aerial mycelium and soluble pigments, nitrate reduction, utilization of sole carbon sources and enzyme activities. The results from the DNA–DNA hybridization studies also support the classification of strain KC-061T as a novel species of the genus Nonomuraea and the name, Nonomuraea thailandensis sp. nov., is proposed.
Description of Nonomuraea thailandensis sp. nov.
Nonomuraea thailandensis (thai.lan.den’ sis. N.L. masc. adj. thailandensis pertaining to Thailand, where the strain was isolated).
Gram-positive and aerobic actinomycete. Substrate mycelia are branched and not fragmented. Depending on the cultivation medium, the color of aerial mycelia is white to pale orange. No aerial mycelia are produced on ISP 4 and 6 media, YS agar and nutrient agar and the substrate mycelia appear pale orange to dark reddish brown. A light brown to deep orange soluble pigment is produced on ISP 2 and 3 media, YS agar and nutrient agar. Growth occurs at 14–38 °C (optimum 20–30 °C) and pH 6 to 11 (optimum pH 7 to 9). The NaCl tolerance for growth is up to 4% (w/v). Utilizes D-glucose, myo-inositol, D-mannitol, melibiose, raffinose, L-rhamnose and D-xylose as the sole carbon sources, but does not utilize sucrose. Reduces nitrate, hydrolyzes casein and starch but does not liquefy gelatin. According to the API ZYM system, the strain shows alkaline phosphatase, leucine arylamidase, α-chymotrypsin, acid phosphatase, naphthol-AS-BI-phosphohydrase and α-glucosidase, β-glucosidase and N-acetyl-β-glucosaminidase activities. Esterase (C4), esterase lipase (C8), valine arylamidase and α-mannosidase of strain are weak activities, and strain shows no activities on cystine arylamidase, trypsin, α-galactosidase and β-galactosidase. The diagnostic amino acid within peptidoglycan is meso-diaminopimelic acid. Whole cell hydrolysates contain galactose, mannose, madurose and ribose. The N-acyl type of muramic acid is acetyl. Diphosphatidylglycerol, phosphatidylmonomethylethanolamine, phosphatidylethanolamine, hydroxy-phosphatidylmonomethylethanolamine, hydroxy-phosphatidylethanolamine, phosphatidylglycerol, ninhydrin positive phosphoglycolipid and unknown phospholipid (PL) are present. The predominant menaquinone is MK-9 (H4) and MK-9 (H0), MK-9 (H2), MK-9 (H6) and MK-10 (H4) are also present. Major fatty acids are iso-C16:0, 10-methyl-C17:0, C17:1 ω8c, C16:1 ω7c/C16:1 ω6c, C16:0, iso-C16:1 G, iso-C15:0, 10-methyl-C16:0 and C16:0 2-OH. The genomic DNA G+C content is 72.4 mol%. The type strain is KC-061T (=JCM 18408T =KCTC 29074T =PCU 327T), which was isolated from soil in Angthong Islands National Park, Surat Thani, Thailand.
Methods
Strain KC-061T was isolated from soil sample collected from Angthong Islands National Park, Surat Thani province, Thailand. The soil sample (1 g) was suspended in distilled water (9 ml) and heated at 55 °C for 5 min followed by a serial dilution method for the isolation using potato starch-glycerol agar (potato starch 1.0%, glycerol 1.0%, K2HPO4 0.2%, (NH4)2SO4 0.2%, MgSO4.7 H2O 0.1%, NaCl 0.1%, CaCO3 0.2%, agar 1.2%, pH 7.0)23 supplemented with nystatin (25 mg l−1) and novobiocin (50 mg l−1). After 2 weeks of incubation at 27 °C, the strain was transferred and purified on yeast extract-malt extract medium (International Streptomyces Project medium 2, ISP 2 medium).24 The strain was maintained on Starch-Yeast extract-Meat extract (SYM agar) (starch 1.0%, N-Z amine 0.3%, yeast extract 0.1%, meat extract 0.1%, CaCO3 0.3%, agar 1.2%, pH 7.0). ‘N. monospora’ PT708T (=TISTR 1910T), N. rhizophila DSM 45382T (=YIM 67092T), N. dietziae (=DSM 44320T) and N. rosea DSM 45177T (=GW 12687T) were used for comparative purposes in the study. In order to observe cultural characteristics, the strain KC-061T, ‘N. monospora’ PT708T, N. rhizophila DSM 45382T, N. dietziae NBRC 14309T and N. rosea DSM 45177T were cultured at 27 °C for 3 weeks on ISP 2, 3, 4, 5, 6 and 7 media,24 YS agar (yeast extract 2.0, starch 1.0, agar 1.5%, pH 7.0) and nutrient agar (Difco). The Color Harmony Manual20 was used to determine the color of aerial and substrate mycelia and soluble pigment. The morphological characteristics were observed by light microscopy and scanning electron microscopy (model JSM-5600, JEOL, Tokyo, Japan), after cultivation on ISP 3 medium at 27 °C for 4 weeks. For scanning electron microscopy investigation, the cultures were fixed with 4% osmium tetraoxide vapor in situ for 16 h at room temperature and then dried at room temperature.25 Physiological characteristics, the temperature range, pH range and NaCl tolerance for growth were determined on ISP 2 medium. Utilization of carbohydrates as sole carbon sources were tested by using ISP 9 medium.26 ISP 4 medium was used for starch hydrolysis test. Nitrate medium (beef extract 0.3%, peptone 0.5%, KNO3 0.1%, pH 7.0) was used for nitrate reduction. Glucose-Peptone-Gelatin medium (glucose 2.0%, peptone 0.5%, gelatin 20%, pH 7.0) was used to test for gelatin liquefaction. Skimmed milk (10%) was used for coagulation and peptonization of milk. The enzyme activities were determined using the API ZYM system (bioMerieux, Lyon, France), according to the manufacturer’s instructions. Biomass for the genotypic study and the chemotaxonomic studies was obtained after cultivation in Yeast Glucose broth (yeast extract 1.0%, glucose 1.0%, pH 7.0) on a rotary shaker at 27 °C for 1 week. Diaminopimelic acid isomers in whole cells were determined by TLC using whole-cell hydrolysis.27 Whole-cell sugar composition was analyzed according to the method of Becker et al.27 Isoprenoid quinones were extracted according to the method of Collins et al.28 and were analyzed by LC/MS (JMS-T 100LP, JEOL) using a CAPCELL PAK C18 UG120 (Shiseido, Tokyo, Japan) with methanol-2-propanol (7:3) as the mobile phase. The N-acyl types of muramic acid were determined by the method of Uchida and Aida.29 Phospholipids in cells were extracted and identified by the method of Minnikin et al.30 The presence of mycolic acids was examined by TLC following Tomiyasu.31 The cellular fatty acid composition analysis was determined by GLC according to the instructions of the Microbial Identification System (MIDI) Sherlock version 6.0 with the RTSBA6 MIDI database as described by Sasser.32
For DNA base composition analysis, chromosomal DNA was prepared following the procedure of Saito and Miura33 and the DNA G+C content was determined by HPLC according to Tamaoka and Komagata.34 DNA–DNA hybridization was performed by the photobiotin-labelling method of Ezaki et al.35 The 16S rRNA gene was amplified using the primers described by Takahashi et al.36 Amplifications were performed in a TaKaRa thermal cycler (Takara Bio Inc., Shiga, Japan) with an initial incubation of 1 min at 95 °C followed by 30 cycles of 1 min at 95 °C, 1 min at 50 °C and 1.5 min at 72 °C, followed by 2 min final extension at 72 °C. The PCR products were sequenced on a DNA sequencer (Applied Biosystems 3130 Genetic Analyzer) using a BigDye Terminator v3.1 cycle Sequencing kit (Applied Biosystems, Foster City, CA, USA), according to the manufacturer’s instructions. The ClustalW2 program was used for multiple alignments with selected sequences for calculating evolutionary distances37 by SeaView version 4.2.38 The phylogenetic trees were constructed based on the neighbor-joining method,39 the maximum-likelihood40 and the maximum-parsimony methods.41 Data were resampled with 1000 bootstrap replications.42 The values for sequence similarities among the most closely related strains were determined using the EzTaxon server.43
References
Zhang, Z., Wang, Y. & Ruan, J. Reclassification of Thermomonospora and Microtetraspora. Int. J. Syst. Bacteriol. 48, 411–422 (1998).
Euzéby, J. P. List of bacterial names with standing in nomenclature: a folder available on the Internet. Int. J. Syst. Bacteriol. 47, 590–592 (1997).
Qin, S. et al. Nonomuraea antimicrobica sp. nov., an endophytic actinomycete isolated from a leaf of Maytenus austroyunnanensis. Int. J. Syst. Evol. Microbiol. 59, 2747–2751 (2009).
Le Roes, M. & Meyers, P. R. Nonomuraea candida sp. nov., a new species from South African soil. Antonie van Leeuwenhoek. 93, 133–139 (2008).
Li, J. et al. Nonomuraea endophytica sp. nov., an endophytic actinomycete isolated from Artemisia annua L. Int. J. Syst. Evol. Microbiol. 61, 757–761 (2011).
Xi, L., Zhang, L., Ruan, J. & Huang, Y. Nonomuraea maritima sp. nov., isolated from coastal sediment. Int. J. Syst. Evol. Microbiol. 61, 2740–2744 (2011).
Zhao, G. -Z. et al. Nonomuraea rhizophila sp. nov., an actinomycete isolated from rhizophere soil. Int. J. Syst. Evol. Microbiol. 61, 2141–2145 (2011).
Kämpfer, P., Busse, H. -J., Tindall, B. J., Nimtz, M. & Grün-Wollny, I. Nonomuraea rosea sp. nov. Int. J. Syst. Evol. Microbiol. 60, 1118–1124 (2010).
Wang, F. et al. Nonomuraea wenchangensis sp. nov., isolated from mangrove rhizosphere soil. Int. J. Syst. Evol. Microbiol. 61, 1304–1308 (2011).
Li, X. et al. Nonomuraea jiangxiensis sp. nov., isolated from acidic soil. Int. J. Syst. Evol. Microbiol. 62, 1409–1413 (2012).
Cao, Y. R., Jin, R. X., Jiang, Y., He, W. X. & Jiang, C. L. Nonomuraea soli sp. nov., an actinomycete isolated from soil. Int. J. Syst. Evol. Microbiol. 62, 1587–1591 (2012).
Goldstein, B. P. et al. A40926, a new glycopeptide antibiotic with anti-Neisseria activity. Antimicrob. Agents Chemother. 31, 1961–1966 (1987).
Gunnarsson, N., Bruheim, P. & Nielsen, J. Production of the glycopeptide antibiotic A40926 by Nonomuraea sp. ATCC 39727: influence of medium composition in batch fermentation. J. Ind. Microbiol. Biotechnol. 30, 150–156 (2003).
Miyanaga, S. et al. Absolute configuration and antitumor activity of myxochelin A produced by Nonomuraea pusilla TP-A0861+. J. Antibiot. 59, 698–703 (2006).
Ayers, S. et al. Anthelmintic Macrolactams from Nonomuraea turkmeniaca MA7381. J. Antibiot. 61, 59–62 (2008).
Terui, Y. et al. WSS2220, a novel cyclic tetrapeptide with a new sulfonoamino acid, exhibits potent and selective inhibitory activity against GlyT1. Tetrahedron Lett. 49, 3067–3070 (2008).
Terui, Y. et al. New cyclic tetrapeptides from Nonomuraea sp. TA-0426 that inhibit glycine transporter type 1 (GlyT1). Bioorg. Med. Chem. Lett. 18, 6321–6323 (2008).
Fleck, F. W., Strauss, D. G., Meyer, J. & Porstendorfer, G. Fermentation, isolation and biological activity of maduramycin: a new antibiotic from Actinomadura rubra. Z. Allg. Mikrobiol. 18, 389–398 (1978).
Paulus, E., Dornberger, K., Werner, W. & Fenske, D. Madurahydroxylactone. Acta Cryst. C 50, 2064–2067 (1994).
Taylor, H. D., Knoche, L. & Grauville, W. C. Color Harmony Manual 4th edn Container Corporation of America: Chicago, (1958).
Nakaew, N., Sungthong, R., Yokota, A. & Lumyong, S. Nonomuraea monospora sp. nov., an antimicrobial and anticancer compound-producing actinomycete isolated from Thai cave soil and emended description of the genus Nonomuraea. Int. J. Syst. Evol. Microbiol. doi:10.1099/ijs.0.035220-0.
Wayne, L. G. et al. International Committee on Systematic Bacteriology. Report of the Ad Hoc Committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Bacteriol. 37, 463–464 (1987).
Tajima, K., Takahashi, Y., Seino, A., Iwai, Y. & Omura, S. Description of two novel species of the genus Kitasatospora Omura et al. 1982, Kitasatospora cineracea sp. nov. and Kitasatospora niigatensis sp. nov. Int. J. Syst. Evol. Microbiol. 51, 1765–1771 (2001).
Shirling, E. B. & Gottlieb, D. Methods for characterization of Streptomyces species. Int. J. Syst. Bacteriol. 16, 313–340 (1966).
Inahashi, Y., Matsumoto, A., Danbara, H., Omura, S. & Takahashi, Y. Phytohabitans suffuscus gen. nov., sp. nov., an actinomycete of the family Micromonosporaceae isolated from plant roots. Int. J. Syst. Evol. Microbiol. 60, 2652–2658 (2010).
Pridham, T. G. & Gottlieb, D. The utilization of carbon compounds by some Actinomycetales as an aid for species determination. J. Bacteriol. 56, 107–114 (1948).
Becker, B., Lechevalier, M. P. & Lechevalier, H. A. Chemical composition of cell-wall preparation from strains of various form-genera of aerobic actinomycetes. Appl. Microbiol. 13, 236–243 (1965).
Collins, M. D., Pirouz, T., Goodfellow, M. & Minnikin, D. E. Distribution of menaquinones in actinomycetes and corynebacteria. J. Gen. Microbiol. 100, 221–230 (1997).
Uchida, K. & Aida, K. Acyl type of bacterial cell wall: its simple identification by a colorimetric method. J. Gen. Appl. Microbiol. 23, 249–260 (1977).
Minnikin, D. E., Patel, P. V., Alshamaony, L. & Goodfellow, M. Polar lipid composition in the classification of Nocardia and related bacteria. Int. J. Syst. Bacteriol. 27, 104–117 (1977).
Tomiyasu, I. Mycolic acid composition and thermally adaptative changes in Nocardia asteroids. J. Bacteriol. 151, 828–837 (1982).
Sasser, M. Identification of bacteria by gas chromatography of cellular fatty acids, MIDI Technical Note 101: North Newwark, DE: MIDI Inc. (1990).
Saito, H. & Miura, K. Preparation of transforming deoxyribonucleic acid by phenol treatement. Biophys. Acta 72, 619–629 (1963).
Tamaoka, J. & Komagata, K. Determination of DNA base composition by reversed- phase high performance liquid chromatography. FEMS Microbiol. Lett. 25, 125–128 (1984).
Ezaki, T., Hashimoto, Y. & Yabuuchi, E. Fluorometric deoxyribonucleic acid-deoxyribonucleic acid hybridization in microdilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains. Int. J. Syst. Bacteriol. 39, 224–229 (1989).
Takahashi, Y. et al. Streptomyces avermectinius sp. nov., an avermectin-producing strain. Int. J. Syst. Evol. Microbiol. 52, 2163–2168 (2002).
Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120 (1980).
Gouy, M., Gascuel, S. & Gascuel, O. SeaView version 4.2: a multiplate form graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27, 221–224 (2010).
Saito, N. & Nei, M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 (1987).
Felsenstein, J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17, 368–379 (1981).
Kluge, A. G. & Farris, F. S. Quantitative phyletics and the evolution of anurans. Syst. Zool. 18, 1–32 (1969).
Felsenstein, J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 39, 783–791 (1985).
Kim, O. S. et al. Introducing EzTaxon-e: a prokaryotic 16S rRNA Gene sequence database with phylotypes that represent uncultured species. Int. J. Syst. Evol. Microbiol. 62, 716–721 (2012).
Acknowledgements
This research was supported by Chulalongkorn University Duthsadiphiphat Scholarship (2008), Chulalongkorn University, Bangkok, Thailand and Kitasato Institute for Life Sciences, Kitasato University, Japan. We would like to thank Dr Mihoko Mori (Kitasato Institute for Life Sciences, Kitasato University) for secondary metabolite study.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Sripreechasak, P., Tanasupawat, S., Suwanborirux, K. et al. Nonomuraea thailandensis sp. nov. isolated from Thai soil. J Antibiot 66, 79–84 (2013). https://doi.org/10.1038/ja.2012.96
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2012.96
Keywords
This article is cited by
-
Nonomuraea indica sp. nov., novel actinomycetes isolated from lime-stone open pit mine, India
The Journal of Antibiotics (2015)
-
Spirotetronate antibiotics with anti-Clostridium activity from Actinomadura sp. 2EPS
World Journal of Microbiology and Biotechnology (2015)
-
Nonomuraea shaanxiensis sp. nov., a novel actinomycete isolated from a soil sample
Antonie van Leeuwenhoek (2014)
-
Nonomuraea guangzhouensis sp. nov., and Nonomuraea harbinensis sp. nov., two novel actinomycetes isolated from soil
Antonie van Leeuwenhoek (2014)