Abstract
The overuse of antibiotics as veterinary feed additives is potentially contributing to a significant reservoir of antibiotic resistance in agricultural farmlands via the application of antibiotic-contaminated manure. Vermicomposting of swine manure using housefly larvae is a promising biotechnology for waste reduction and control of antibiotic pollution. To determine how vermicomposting influences antibiotic resistance traits in swine manure, we explored the resistome and associated bacterial community dynamics during larvae gut transit over 6 days of treatment. In total, 94 out of 158 antibiotic resistance genes (ARGs) were significantly attenuated (by 85%), while 23 were significantly enriched (3.9-fold) following vermicomposting. The manure-borne bacterial community showed a decrease in the relative abundance of Bacteroidetes, and an increase in Proteobacteria, specifically Ignatzschineria, following gut transit. ARG attenuation was significantly correlated with changes in microbial community succession, especially reduction in Clostridiales and Bacteroidales. Six genomes were assembled from the manure, vermicompost (final product) and gut samples, including Pseudomonas, Providencia, Enterococcus, Bacteroides and Alcanivorax. Transposon-linked ARGs were more abundant in gut-associated bacteria compared with those from manure and vermicompost. Further, ARG-transposon gene cassettes had a high degree of synteny between metagenomic assemblies from gut and vermicompost samples, highlighting the significant contribution of gut microbiota through horizontal gene transfer to the resistome of vermicompost. In conclusion, the larvae gut microbiome significantly influences manure-borne community succession and the antibiotic resistome during animal manure processing.
Similar content being viewed by others
Introduction
China is the largest producer and consumer of antibiotics in the world, and more than 46% of antibiotics are used for growth augmentation and disease control in livestock industries (Hvistendahl, 2012; Zhu et al., 2013). The prevalence of concentrated swine feed operations in China also creates the largest global volume of swine manure (Wang et al., 2006). Animals only absorb a small concentration of consumed antibiotics, so swine manure becomes a major source of environmental antibiotic pollution (Alcock et al., 1999; Zhu et al., 2013), and unprocessed manure is therefore a significant potential reservoir of antibiotic resistance in environmental microbiota (Smillie et al., 2011; Forsberg et al., 2012).
Traditional waste composting by aerobic digestion leads to a significant increase in antibiotic resistance genes (ARGs) in the environment (Zhu et al., 2013; Su et al., 2015), which is likely due to the persistence and mobility of manure-borne bacterial strains, with certain taxa preferentially hosting resistance genes (Ghosh and LaPara, 2007; Looft et al., 2012). These resistant strains may also act as a source for the horizontal transfer of resistance genes into environmental bacteria (Enne et al., 2001; Gaze et al., 2011), which can be widely distributed (Li et al., 2015a). Therefore, we urgently need to evaluate biotechnologies that reduce the proliferation of resistance genes in animal waste.
Manure management can be augmented by the activity of earthworms, Naididae (previously Tubificidae), housefly larvae, black-soldier fly larvae and other species. This form of bioconversion can be traced back to the 1970–1980s (Calvert et al., 1970; Neuhauser et al., 1980; Hartenstein and Hartenstein, 1981). However, bioconversion using insects is still not popular owing to the handling requirements necessary to rear these animals (Wang et al., 2013). Recently, swine manure vermicomposting aided by housefly larvae (Musca domestica) has reached full-scale operation in China (Zhang et al., 2012; Wang et al., 2013; Zhang et al., 2014). Larvae conversion of swine manure results in a significant decrease in waste volume and antibiotic residues (Zhang et al., 2012; Zhang et al., 2014), as well as a significant decrease in microbial biomass and activity (Zhang et al., 2012; Wang et al., 2013). However, more interesting is the potential for manure-associated bacteria and larvae gut-associated bacteria to interact, which could have profound ecological repercussions, changing the selection pressures in this environment, and hence the persistence of ARGs.
We hypothesize that larvae gut transit influences bacterial community composition and the selection for ARGs in swine manure. To test this hypothesis, ARG dynamics during vermicomposting were quantified using qPCR, and microbial community structure and function were assessed using both 16S rRNA amplicon sequencing and de novo assembly of metagenomic data. Associations between ARG abundance and community dynamics were calculated with local similarity analysis, and Bayesian approaches were used to determine the sources of bacteria in larvae-treated samples (from either raw manure or gut microbiota). Further, we interpreted the selection pressure and rate of evolution for bacterial genomes and the potential association of mobile genetic elements with ARGs in each environment. We demonstrated that the ingested manure-borne bacteria were affected by larvae gut transit, through eco-evolutionary pressures that altered community structure and the resistome.
Materials and methods
Manure sampling and DNA extraction
Samples were collected from greenhouse-assisted larvae bioreactors (Supplementary Figure S1) in a full-scale swine manure vermicomposting operation (30°49′47.02″N, 120°39′22.12″E). A continuous-feeding strategy was applied to treat daily-generated raw manure (approx. 25 m3 of fresh manure per day; Supplementary Figure S2). Raw manure was consistently obtained from a neighboring finishing swine farm, which produces approximately 35 000 tons of fresh manure annually. Tetracyclines were used for in-feed antibiotics, while sulfonamides, aminoglycosides and quinolones were used for therapy in this farm. The exact dosage of each antibiotic applied in swine feed was not determined; however, the doses were reported to be consistent over time. Raw manure samples (S0d) from this swine farm were first randomly collected from larvae bioreactors in May 2013. Raw manure under vermicomposting from day 1 to day 6 was then collected (that is, larvae-treated samples, S1d, S2d, S3d, S4d, S5d and S6d) using longitudinal time-course sampling protocols. S6d represents the vermicompost (final product). The early (that is, 12–20 h after fly oviposition) or late instar (that is, 24–48 h before pupation) larvae were simultaneously collected for each day (that is, larvae gut samples, L1d, L2d, L3d, L4d, L5d and L6d). At the same time, samples as C0d, C1d, C3d and C6d collected from naturally piled-up composting with no larvae input (that is, traditional composting) served as controls in this study.
Total DNA was extracted using the PowerSoil DNA Isolation Kit (MO BIO Laboratories, Carlsbad, CA, USA) following the manufacturer’s protocols, and was normalized to the equal concentrations before downstream processing. Before larvae gut DNA extraction, the collected early or late instar larvae were starved for 8 h to empty their ingested contents, and then about 600 larvae were anatomized for each tube of total DNA extraction from larvae’s midgut and hindgut (Supplementary Figure S2B).
High-capacity qPCR for ARGs
A total of 45 DNA samples (Supplementary Table S1) were analyzed for 285 ARGs on a Wafergen SmartChip nanowell real-time qPCR platform, with reaction protocols described by Wang et al. (2014). Three technical replicates were performed for each replicate. A threshold cycle (Ct) of 31 was used as the detection limit. When multiple primer sets targeted the same gene, it was only counted as the detection of single unique ARG (Supplementary Table S2). A mathematical model described by Pfaffl (2001) was applied to determine the relative abundance (gene copies normalized to 16S rRNA gene) of ARGs for each sample. Fold changes in gene abundance were further calculated by comparing all other samples with C0d as a reference sample (Supplementary Table S4). In calculating fold changes, if there was no amplification for ARGs, the detection limit Ct (31) was used. These detected ARGs were classified based on the mechanism of resistance, and the antibiotics to which they confer resistance.
16S rRNA amplicon sequencing, data processing and analysis
A total of 51 DNA samples (Supplementary Table S1) were sequenced. The V4 region of bacterial 16S rRNA was amplified using a universal primer set 515F/806R on a 150 bp pair-end Illumina Hiseq (San Diego, CA, USA) sequencer in Argonne National Lab. The raw reads have been deposited into the DDBJ Sequence Read Archive (SRA) database (Accession number: SRP050167). The QIIME pipelines v1.8.0 (Caporaso et al., 2010) were used for pre-processing of the raw reads from a total of 51 amplicon libraries. After quality filtering and demultiplexing, replicates of C6d.3, L5d.2 and S4d.1 contained <100 reads and were therefore not used in downstream analyses. USEARCH v6.1 was used to identify chimeras (about 5.4%). We obtained on average 54 250 high-quality reads per replicate, with an average length of 253 bp. Using a closed-reference operational taxonomic unit (OTU) picking strategy, 6436 OTUs were identified at a 97% similarity threshold using uclust v1.2.22q to cluster against Greengenes reference database (13_8 release) (McDonald et al., 2012). Taxonomy was achieved using RDP Classifier (Cole et al., 2009) to assign against the same reference.
Before further analysis, the reads were rarefied or normalized to correct for differences in sequencing depth if needed. Alpha and beta diversity of bacterial community structure was analyzed using QIIME (http://qiime.org.html). SourceTracker (http://qiime.org/tutorial/source_tracking.html) (Knights et al., 2011) was used to profile the potential sources of microbial communities in a set of input samples (that is, manure sample) shaped by larvae gut-associated microbiota. Rare taxa presented in less than five samples were removed. Manure under vermicomposting from day 1 (S1d) to day 6 (S6d) were considered as a sink for each day, and raw manure and larvae gut samples were considered as two major known sources, respectively. ‘Unknown’ assignments represent reads with no mapping to our input ‘sources’.
Oligotyping was used to determine the sub-species level assignment for Ignatzschineria, Bacteroides, Alcaligenes, Prevotella, Acinetobacter, Anaerococcus, Providencia and Pseudomonas following the pipeline version 0.96 (available from http://oligotyping.org) to determine whether gut also reshaped communities at finer resolution. We set M to equal the average number of reads per sample divided by 1000, s to match the number of replicates (s=3) and c to the number of maximum entropy locations. Such a configuration will eliminate oligotypes that appear in fewer than s samples and with very low substantive abundance (Eren et al., 2013). To assign each oligotype to its taxonomy, the representative sequence (the most abundant unique one) for each oligotype was searched against a 16S rRNA sequence (Bacteria and Archaea) database using BLASTn (http://www.ncbi.nlm.nih.gov/) with default parameters. The hits with the most similarity for each oligotype were reported. When similarity scores less than 97%, it was assigned as ‘unclassified’.
The functional profiling was inferred from 16S rRNA marker gene sequences using PICRUSt (Langille et al., 2013). For all studied samples, NSTI was kept less than 0.10 to make sure the accuracy of metagenome inference. The identified KOs were then binned into functional categories under a given subsystem hierarchy level II and level III derived from KEGG modules.
Local similarity analysis between taxa and ARGs
Local similarity analysis (Xia et al., 2011) was performed for accessing the correlations between relative abundance of OTUs or annotated taxa and ARGs detected in manure during vermicomposting (from day 0 to day 6). Local similarity score (LS score) and P-value for each correlated pair was set as >0 and <0.01, respectively. For original constructed relationships, the basic network parameters were obtained. To filter the data for reduced network complexity, OTUs and taxa were further filtered to have a relative abundance no less than 0.1% of the total. Networks were visualized using Cytoscape 3.3.
Shotgun metagenomics of manure or larvae gut-associated samples
Sequencing and quality control
A total of nine samples (C0d, S0d, L1d–L6d and S6d, Supplementary Table S1) were selected for shotgun metagenome sequencing. Using Nextra XT protocols (Illumina), individual libraries were sequenced on the Miseq platform. Paired end reads were quality-trimmed using nesoni pipeline (https://github.com/Victorian-Bioinformatics-Consortium/nesoni), with the parameters set at: minimum length=75, quality cutoff=30, adapter trimming yes and ambiguous bases=0. Quality-trimmed reads (larvae samples) that mapped to the housefly genome (GenBank accession number AQPM00000000.1 and NCBI assembly accession GCA_000371365.1) were identified and removed using BWA aligner (Li and Durbin, 2009). Reference genome mapping revealed that 30% of the larvae sample reads were originated from the housefly DNA. The sequences reported in this study were deposited in MG-RAST, http://metagenomics.anl.gov/ (project ID 1582, metagenomes 4671865.3, 4671866.3, 4671872.3, 4671874.3- 4671877.3, 4671879.3 and 4671880.3).
Metagenome assembly, annotation and de novo genome reconstruction
Preliminary metagenome assemblies were generated for each sample using MEGAHIT (Li et al., 2015b) with parameters set as follows: –min-k=41 –max-k=51. To reconstruct de novo population genomes, a composite metagenome assembly was generated for larvae gut samples (n=6), raw manure (C0d and S0d) and vermicompost (S6d) using MEGAHIT set at same parameters. MetaBAT pipeline (https://bitbucket.org/berkeleylab/metabat) was used (mode=—specific) for binning genomes from composite metagenome assembly (contigs>3 kb). Percentage completeness estimations of the reconstructed genomes were performed using CheckM (Parks et al., 2015). Genomes (n=6) greater than 80% completion were used for detailed downstream analyses. Draft genomes were checked for sequence decontamination using ProDeGe pipeline (https://prodege.jgi-psf.org/). Metagenomic contigs were annotated at enzyme and pathway level using prokka pipeline (Seemann, 2014). Bacterial transposase and integrase sequences were downloaded from UniProt database (http://www.uniprot.org/) and were formatted into individual BLAST databases, and used to annotate metagenomic reads (BLASTX; e-value=10−3). Taxonomy was assigned to the reads associated with the integrase and transposase sequences using Kraken (Wood and Salzberg, 2014).
Functional annotation and natural selection analysis of reconstructed genomes
Protein coding genes were predicted across each genome using FragGeneScan (Rho et al., 2010). Functional status was assigned to each gene using KAAS server (Moriya et al., 2007). Automated genome annotations were performed using RAST server (Aziz et al., 2008). For the comparison of functional similarity between 16S rRNA and metagenomic analysis, multi-group and two-group analysis were performed using analysis of variance and Welch’s t-test with false discovery rate correction, respectively. To predict the coding sequence for antibiotic resistance, assemblies were screened against Antibiotic Resistance Genes Database (ARDB) (Liu and Pop, 2009). Reciprocal Smallest Distance algorithm (Wall and DeLuca, 2007) was used to predict pairwise orthologous genes between reconstructed genomes and their closest phylogenetic neighbor (predicted using RAST functional score cutoff).
Results
Dynamics of ARGs in manure during larvae gut transit
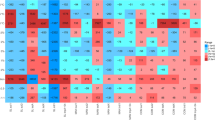
We detected 158 unique ARGs among samples C0d, C3d, C6d and S0d-S6d, which encompass three major resistance mechanisms: antibiotic deactivation, efflux pumps and cellular protection (Supplementary Figure S3). Genes conferring resistance to aminoglycoside, (flor)/(chlor)(am)/phenicol, Macrolide-Lincosamide-Streptogramin B resistance (MLSB), tetracycline, as well as sulfonamide were all detected in raw manure; the top 10 most abundant genes for raw manure were aadA1, strB, aadA-02, aadA-01, tetQ, tetM, ermF, cfr and floR, which represented a core antibiotic resistome for these samples (Figure 1a, Supplementary Tables S3 and S4). After 6 days of larvae treatment, 94 unique genes were attenuated (P<0.05) in vermicompost by 85%, when compared with raw manure (Figure 1b), which mainly encoded for antibiotic deactivation and cellular protection, conferring resistance to beta_lactamase, MLSB and vancomycin. Meanwhile, 23 unique ARGs were enriched (P<0.05) by 3.9-fold (Figure 1c). When S6d was compared with C6d (Supplementary Figure S4), the patterns were similar to those compared with C0d (Figure 1). These enriched genes (such as aminoglycoside- and sulfonamide-resistant genes) have been frequently reported in mobile elements (Supplementary Table S5), indicating they could persist by horizontal gene transfer.
Attenuation or enrichment of ARGs. (a) Profiling of ARG abundance for all observed samples (with 12 replicates for C0d and S6d and 3 for all other samples; 6 S0d samples were marked as C0d here, because they are biologically the same). Each row is the relative gene abundance from a single primer set, with only those that showed significant (P<0.05) changes among samples determined by analysis of variance plus Duncan tests represented. Red indicates greater correlation, blue indicates less correlation, and green represents median values. ARGs highlighted in yellow are specified in Supplementary Table S5; and all datasets are outlined in Supplementary Table S3. (b) The descriptive statistics for attenuated resistance genes (the relative abundance, expressed as S6d/C0d when <1). The box plots denote 25th to 75th percentile; horizontal line, median; and whiskers, 10th and 90th percentile. Symbols along x axis indicate genes conferring resistance to aminoglycoside (Am), beta_lactamase (Be), (flor)/(chlor)/(am)phenicol (Ph), (flouro)quinilone (Qu), MLSB (Ml), Multidrug (Mu), Tetracylines (Te), Vancomycin (Va) and Sulfonamide (Su). (c) The descriptive statistics for enriched resistance genes (S6d/C0d >1). Fold change values are considered significant only if the range created by standard deviations of the average are entirely <1 (attenuated) or entirely>1 (enriched) (see Supplementary Table S4 for the datasets).
Reshaping of bacterial community composition over larvae gut transit
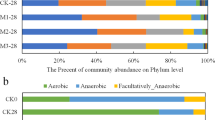
Bacterial species richness significantly decreased during vermicomposting compared with raw manure and the traditional composting samples (without larvae). The larvae gut-associated microbiota had the lowest diversity (Supplementary Figures S5 and S6), and also showed the lowest variance in community structure (beta diversity) between samples from day 1 to day 6 (Figure 2a). Vermicomposting samples were highly divergent and formed distinct clusters: the first few days of vermicomposting were closest in similarity to raw manure, while at the end of the process, the vermicompost is more similar to the gut-associated microbiota (Figures 2a and b). Vermicompost was significantly enriched in Proteobacteria and was depauperate for Bacteroidetes (P<0.05) compared with raw manure. Proteobacteria dominated in the larvae gut (92% of all sequences), which comprised mostly the genera Ignatzschineria (50%), Providencia (21%) and Pseudomonas (12%, Figure 2b and Supplementary Table S6). However, Providencia was not abundant in the vermicompost, which was dominated by Ignatzschineria (30%) and Pseudomonas (6.3%). Ignatzschineria was 28 × more abundant, and Pseudomonas was twice as abundant in vermicompost compared with raw manure (Figure 2b). By comparison, Bacteroides and Prevotella, both known to dominate in the swine gut, were extremely rare in the vermicompost (Figure 2b and Supplementary Table S6).
Bacterial community structure from 16S rRNA analysis, and functional predictions using PICRUST. (a) PCoA plots based on weighted UniFrac distances of samples. Red triangle=manure samples under vermicomposting (red triangle); green circle=manure samples under traditional composting without larvae input; blue square=larvae gut samples. The average distances for each category by connecting all points with minimal total length divided by the total numbers of these involved samples are further shown. (b) Relative abundance (%) of taxa at the genus level, clustered using UPGMA on the weighted UniFrac distances. Taxa with <1% of reads were merged together as ‘others’; while ‘unknown’ represents unclassified taxa at the genus level (see Supplementary Table S6). (c) PICRUST predictions using KEGG subsystem level II. Raw manure (C0d+S0d), larvae gut (L1d to L6d) and vermicompost (S6d) are compared using non-parametric Kruskal–Wallis test with Dunn’s multiple comparison. Only functions with >1% abundance are shown. Red on y axis indicates increased (P<0.05) functional abundance in vermicompost compared with raw manure (see Supplementary Table S13).
Linking ARGs with larvae-associated microbiome
Using local similarity analysis, some of the attenuated or enriched ARGs were found to correlate significantly with the dynamics of certain bacterial clusters (that is, taxa or OTUs) (Figure 3 and Supplementary Table S7). The attenuated ARGs-related network was more complex than the enriched one: an average of 30.3 nodes were correlated with each attenuated ARG (Figure 3a) compared with only 5.4 nodes with the enriched (Figure 3b).
Local similarity analyses of the potential linkages between the dynamics of ARGs and bacterial taxa/OTUs during vermicomposting. Blue octagons represent taxon at phylum, class, order, family or genus level; orange circles represent OTUs (97% similarity threshold); and green diamonds represent ARGs. Node size represents average relative abundance across replicates. Edge color represents local similarity score, and weight represents P-value, where a lower P-value will have a wider edge. (a) Subnetwork organized between significantly attenuated ARGs and taxa/OTUs. (b) Subnetwork organized between significantly enriched ARGs and taxa/OTUs. (c) Subnetwork organized between significantly attenuated ARGs and family members belonging to the order Clostridiales. (d) Subnetwork organized between significantly attenuated ARGs and family members belonging to the order Bacteroidales. (e) Subnetwork organized between significantly attenuated ARGs and OTUs/taxa belonging to the family Bacteroidaceae. To reduce the network complexity, pairs illustrated here were filtered with a significant P-value<0.005. The basic network parameters for original network diagrams (before data filtration) are shown in an inserted table. Also see Supplementary Table S7 for taxonomic information for each OTU in network.
ARGs were associated with nodes of different taxonomic ranks from Phylum to OTU (97% similarity threshold) for the attenuated network (Figure 3a). For example, ermJ/ermD, vgaA-01, blaGES and vanXB were only correlated with Bacteroidetes (P<0.01), and cfr and vatE-01 were only correlated with the genus Acinetobacter and the family Prevotellaceae (P<0.01), respectively. Bacteria correlated with attenuated ARGs mainly belonged to the order Clostridiales and Bacteroidales, the dominant taxa in raw manure samples. By exploring the subnetworks of attenuated ARGs with the orders Clostridiales (Figure 3c) and Bacteroidales (Figure 3d), we found that most families of both orders have similar antibiotic resistance profiles, except for the family Prevotellaceae in Bacteroidales and the family Tissierellaceae in Clostridiales. Both Prevotellaceae and Tissierellaceae correlated with ARGs that were likely to be located in mobile elements. The subnetwork of Bacteroidaceae, the most abundant family of Bacteroidales, showed a similar scenario (Figure 3e). This suggests that taxa from the same lineages may have similar ARG profiles, suggesting taxonomic conservation. For most of the attenuated ARGs that were not located in mobile elements, their correlations with certain bacterial clusters (taxa/OTUs) were more likely to be linear relationships. Enriched ARGs, mphA, aadA2, aadA5-2, aadA9-02 and tetG-01 (Figure 3b, big cluster), as well as cmlA1and tetR-02 (Figure 3b; small cluster), were mostly correlated with genera or OTU (97% similarity) level clusters (Figure 3,Supplementary Table S7), which mainly belonged to the family Alcaligenaceae, [Weeksellaceae] and the order Bacillales. Their abundances increased distinctly during vermicomposting from 0.18%, 0.38% and 0.64% to 13%, 26% and 4.5%, respectively. These taxa-linked ARGs were more likely to be located in mobile elements, compared with attenuated ARGs (Figure 3, Supplementary Table S5). Pseudomonas was also found to correlate with enriched ARGs (ampC-07, tet-x and tetL-02; Figure 3b). OTU 109756 in the family Pseudomonadaceae was positively correlated with mphA, aadA2, aadA5-2, aadA9-02 and tetG-01 (Figure 3b, big cluster). Genus Ignatzschineria, the most abundant taxon post-vermicomposting, was only correlated with aacC1.
Determining the potential sources of taxa found in vermicompost
SourceTracker, a Bayesian classifier approach, was used to characterize the origin of taxa in treated samples from day 1 to day 6, respectively (Figure 4a). On day 1, 91% of vermicompost OTUs originated from raw manure, and only 1.4% from larvae gut. By day 2, only 63% of taxa came from manure, and by day 6, this had dropped to 15%. Clostridiales and Bacteroidales dominated raw manure (26% and 48%, respectively), and on day 1 of vermicomposting (16% and 27%, respectively), but by day 6, they only comprised 1.7% and 1.2% of the total bacterial community, respectively. The larvae gut microbiota increased as a source to vermicompost, and taxa of ‘unknown’ origin became a dominant contributor on day 6 (Figure 4a). We examined OTUs of ‘unknown’ origin exclusively found in vermicompost, but absent in raw manure and gut samples (Figure 4b). OTUs belonging to family Alcaligenaceae, genus Pseudidiomarina, Erysipelothrix and order Bacillales, had greater abundance in vermicompost (Supplementary Table S8) compared with raw manure and gut samples. Succession of sub-species level taxa under treatment was determined using oligotyping (Figure 4c and Supplementary Table S6). For example, using oligotyping, Ignatzschineria comprised two oligotypes, which belonged exclusively to I. indica and I. ureiclastica (Figure 4c and Supplementary Table S6). We also identified six oligotypes for Pseudomonas. Two of these oligotypes, ‘AA’ and ‘AC’, demonstrated contrasting trends over the 6 days (Figure 4c). These two oligotypes were identified as sub-species of Pseudomonas brenneri and Pseudomonas formosensis (100% nucleotide sequencing identity using BLAST, Supplementary Table S6). This suggests that fine-scale changes in population dynamics under vermicomposting may need to be considered for future analysis.
Larvae gut reshapes bacterial community compositions in manure samples. (a) Source Tracking of microbial communities in manure under vermicomposting from day 1 (S1d) to day 6 (S6d). ‘Unknown’ are taxa that could not be significantly soured to either raw manure or larvae gut. (b) Exclusive and shared OTUs (non-singleton OTUs, based on 97% reads similarity) between raw manure (C0d and S0d), larvae gut (L1d to L6d) and vermicompost (S6d), with number representing OTUs found in each segment (see Supplementary Table S8). (c) Oligotyping of Ignatzschineria and Pseudomonas. The dynamics for each identified oligotype are shown from 1 day (S1d) to 6 day (S6d) under vermicomposting (see Supplementary Table S6).
The antibiotic resistome of swine manure revealed by reconstructed genomes
Nine metagenomes (raw manure=2, larvae gut=6 and vermicompost=1) were sequenced, producing between 14 and 32 million quality-trimmed reads each. The microbial community profile determined by metagenomic analysis was similar to that determined by 16S rRNA amplicon analysis at both phylum and genus levels, except that Ignatzschineria was absent from the metagenomic study (Supplementary Figure S7) as no genome was available to annotate reads against. Metagenomic assembly was performed to examine the genomes of enriched microbial taxa only. In total, 22 genome bins were assembled, but 16 of these were <80% complete, and were therefore excluded from downstream analysis. Three genome bins, Pseudomonas, Enterococcus and Providencia, were reconstructed from gut samples. In addition, Alcanivorax and Bacteroides genomes were reassembled from the vermicompost and raw manure samples, respectively (Table 1). ARGs were more abundant in genomes reassembled from vermicompost (n=14) compared with manure (n=9) and gut (n=5) samples (Supplementary Table S9). Complete functional annotation (enzyme and pathway level) of reconstructed genomes is provided in Supplementary Tables S10 and S11.
Gene-centric metagenomic analysis revealed significant (P<0.05, analysis of variance; false discovery rate-corrected) differences in total community metabolism across vermicompost, manure and gut samples (Supplementary Table S12). However, two-group analysis further revealed significantly similar (R2=0.8; P<0.05; Welch's t-test; false discovery rate-corrected) community metabolism profiles across the vermicompost and gut (Supplementary Table S12). Discounting core metabolism (ko01100; translation, transcription and energy metabolism), the biosynthesis of secondary metabolites (ko01060; vermicompost=7.50 and gut=7.66, raw manure=3.40) and biosynthesis of antibiotics (ko01130; vermicompost=5.33 and gut=5.23, raw manure=2.00) were significantly more abundant in vermicompost and gut samples compared with manure samples (Supplementary Table S12). To extend the functional analysis, we generated predicted metagenomic profiles using PICRUSt (Supplementary Table S13). For samples where metagenomic data were available (n=9), PICRUSt predictions for the relative abundance of KEGG pathways were significantly similar (P<0.05; Welch’s two-group t-test) to those represented in the gene-centric annotation of metagenomic protein-coding genes. Vermicompost samples were significantly enriched for functions involved in cell motility, and membrane transport and metabolism of, for example, lipid and xenobiotics, compared with raw manure (analysis of variance; P<0.05; Figure 2c). Vermicompost functional potential also closely paralleled that of the larvae gut (Figure 2c and Supplementary Figure S8), indicating that the gut-associated microbial community significantly influenced the functional composition of manure.
Metagenomic reads and assembled contigs were mapped against a custom mobile genetic elements database consisting of 149 916 transposase and 56 953 integrase protein sequences. There were no significant differences in the relative abundance of integrases (gut=491, manure=235 and vermicompost=329) or transposases (gut=521, manure=352 and vermicompost=481) between manure, vermicompost and gut samples. However, metagenomic contigs that contained both an ARG and a linked transposon (ARG-transposon cassettes) were more abundant in the larvae gut samples (manure=4, vermicompost=7 and gut=18); 5 ARG-integron cassettes were also identified in guts compared with 2 in raw manure samples. The cmlA1, aadA1 and aadA2 genes were across larvae gut integron gene cassettes, especially associated with class 1 integrons, whereas sul1 gene was identified across manure integron gene cassettes. ARG-transposon cassettes, enriched in Bacteroides, mainly contained tetracycline resistance alongside transposons in raw manure samples. Cassettes with taxonomic similarity to Pseudomonas, Acinetobacter and Providencia contained aminoglycoside (COG2746; vermicompost=3, gut=5 and manure=2), tetracycline (PRK14751; vermicompost=1, gut=5), multidrug (TIGR00880) and hygromycin (vermicompost=1, gut=1) resistance genes mainly found in gut and vermicompost. Moreover, we found two nearly identical (>98% nucleotide identity) cassettes in vermicompost and gut samples showing synteny for the transposon and ARG sequences (aph-I; aminoglycoside phosphotransferase and hpt; hygromycin phosphotransferase; Supplementary Figure S9). The greater abundance of ARG-transposon cassettes in the gut-associated microbiome, and the differential abundance of ARGs in the reconstructed genomes, suggests that ARGs have greater horizontal gene transfer potential in the larvae gut samples, compared with the manure environment.
Discussion
The microbiome of raw manure was significantly altered when passed through the gut of housefly larvae, leading to the enrichment of certain taxa originating from the gut, which has a greater association of ARGs with mobile genetic elements.
The benefit of using saprophagous macroinvertebrates (worms or insects) to reduce waste streams may be in part related to the activity of their gut-associated bacteria and the gut environments (Liu et al., 2008; Dominguez et al., 2010; Zhang et al., 2012). The microbiota of earthworms (Egert et al., 2004), termites (Warnecke et al., 2007), coleopteran/dipteran larvae (Egert et al., 2003) and housefly larvae are thought to decompose organic substrates differently (Su et al., 2010). Earthworm or termite guts generally showed higher bacterial diversity than their surroundings (Bernard et al., 2012). In contrast, the housefly larvae gut possessed a less diverse microbiota, lower species richness and distinctly different taxonomic and functional compositions compared with ingested manure (Figure 2). The gastrointestinal tract of the larvae has very low pH and secretes diverse proteolytic enzymes and inhibitory substances (Lemos and Terra, 1991; Mumcuoglu et al., 2001; Blahovec et al., 2006), which likely exerts a strong selective pressure on gut microbiota and the ingested manure-borne microbiome (Supplementary Figure S10). A large number of bacteria will likely be killed and digested during Musca domestica gut transit, and the resulting feces contain only small numbers of species (Mumcuoglu et al., 2001), which may explain the overall attenuation of the resistome in our study (Figure 3). The most abundant manure-borne taxa, Clostridiales (26% in raw manure) and Bacteroidales (48% in raw manure), were significantly attenuated during the gut transit; the reduction in these two taxa also correlated with the most significant attenuation in ARGs, although this could be a statistical anomaly. A striking finding is that the most attenuated ARGs were not associated with mobile elements (Supplementary Table S5), and thus are unlikely to be transferred from dead to living bacterial cells. The existence of DNases under gut conditions may further degrade DNA, which can serve as a nutrient source for gut microbiota (Mumcuoglu et al., 2001; Blahovec et al., 2006).
The larvae gut itself harbored a unique microenvironment where the genera Ignatzschineria, Providencia, Pseudomonas and Morganella dominated (Supplementary Table S6). These taxa are always detected in gastrointestinal tract and stool of M. domestica (Su et al., 2010). Other genera, such as Providencia can produce urease, which neutralizes acidic digestive substances and protect bacteria from enzymatic hydrolysis (Ozogul, 2004; Su et al., 2010). Morganella can synthesize high histidine decarboxylase and tolerate low pH environments and so are able to colonize the larvae gut (Ozogul, 2004).
The larvae gut selected for certain species (Ignatzschineria, Pseudomonas and Alcaligenes), which dominated in vermicompost after treatment. By using SourceTracker analysis (Figure 4 and Supplementary Table S6), we showed that these enriched genera were of gut microbiota origin and/or further selected by surrounding environments (such as Alcaligenes, the OTUs of which were exclusively dominant in vermicompost, Supplementary Table S8). The increase in OTUs of unknown origin by the last day of the vermicomposting treatment suggests that other sources can predominate. These taxa were mostly aerobic or facultatively aerobic, and likely originated from other environments, maybe arriving via insects or air currents, or even on the skin or clothing of workers who maintain the vermicompost. However, it is also possible that they were below the level of detection in the manure and gut samples, and suddenly increased in abundance because of increased oxygen penetration in vermicompost. The enriched Ignatzschineria in vermicompost was mainly from gut microbiota, indicating certain bacteria residing in treated samples can be traced back to the larvae gut. Meanwhile, vermicomposting greatly altered the physicochemical properties of the raw manure (Zhang et al., 2012; Wang et al., 2013; Zhang et al., 2014), which led to a portion of enriched bacteria matching neither raw manure nor gut microbiota. These enriched taxa mainly belong to aerobic or facultative anaerobic bacteria, which explain their survival and propagation in oxygen-filled vermicompost (Mendes et al., 2014). Overall, bacterial community in vermicompost displayed a transitional, intermediate status between the raw manure and gut samples (Figure 2a).
Confirmed by our metagenomic data, some ARGs that were frequently reported in mobile genetic elements (Supplementary Table S5), such as conjugative plasmids, integrons and transposons (Zhu et al., 2013; Su et al., 2014) were not ablated by vermicomposting treatment. Consistently, there was also strong evidence showing that the housefly gut environment permits greater horizontal transfer of the resistome among Escherichia coli strains via plasmid transfer or phage transduction (Petridis et al., 2006). The key to gene transfer-driven bacterial evolution and adaptation involves a variety of mobile genetic elements (Aminov, 2011). In our study, the larvae gut harbored more ARG-integron and ARG-transposon cassettes compared with raw manure. Some ingested ARGs can persist through the gut environment as a result of mobile genetic element assisted transfer (Petridis et al., 2006; Aminov, 2011). Our studied manure was an important reservoir of aadA gene cassettes, and bacteria-bearing aadA can be easily transferred from manure to vermicompost (Binh et al., 2009). PCR analysis demonstrated that larval feces contained aadA genes at considerable concentrations (up to 109 copies per gram of feces) (Brinkmann and Tebbe, 2007), which also suggests that these genes may persist through horizontal gene transfer. More than 100 different gene cassettes have been reported to carry ARGs (Mazel, 2006; Boucher et al., 2007). Among these, aadA gene cassettes (aadA1, aadA2, aadA9) were enriched 24-fold after vermicomposting. Other genes enriched after treatment, such as cmlA1, mphA and blaCTX-M, are also association with class 1 integrons (Naas et al., 2001; Poole et al., 2006). Our metagenomic assembly confirmed this association with integron gene cassettes. Some ARGs, such as ampC and blaCTX-M, were not detected in raw manure, but they occurred in vermicompost over time (S1d for ampC and blaCTX-M; Supplementary Table S3). One possible explanation is that the bioreactors for vermicomposting are open systems and as such, new ARGs could be introduced from external environments, which is supported by the SourceTracker analysis (Figure 4a). Alternatively, these genes were below the limit of detection in manure.
Attenuated ARGs showed a much greater degree of connectivity with bacterial taxa in local similarity analysis than enriched ARGs (Figure 3). This implies that a reduction in bacterial diversity accompanied the elimination of certain ARGs, which were more likely associated with the ablated manure-borne taxa. However, the composting of household organic waste and sewage sludge can select for antibiotic resistant microorganisms, with the most diverse multi-resistant profiles found in Pseudomonas and Ochrohactrum (Heck et al., 2015). Similarly, in compost-amended soils, Pseudomonas was persistent and appears to have acquired multidrug resistance through horizontal gene transfer (Riber et al., 2014). Pseudomonas contained cassettes harboring genes conferring resistance to aminoglycoside and tetracycline (Table 1). In addition, the enriched genes which conferred resistance to MLSB and (flor)/(chlor)/(am)phenicol showed close correlations with Bacillales, Alcaligenaceae and [Weeksellaceae] (Figure 3). Species in Bacillales can excrete cellulase and polyphenoloxidase, which are thermo-tolerant, so enable compost maturity; they can survive the harsh environment and become effective recolonizers. (Mayende et al., 2006). Alcaligenaceae and [Weeksellaceae] also underwent a growth spurt and dominated in the vermicompost of day 6. With the occurrence of horizontal gene transfer during the vermicomposting, these enriched taxa may act as the recipients of ARGs (Boucher et al., 2007; Ramsden et al., 2010).
The long-term goal of manure management is to realize waste volume reduction and maintain the manure as a fertilizer (Wang et al., 2013), but it is also necessary to decrease potential environmental risks associated with antibiotic resistance. We conclude that vermicomposting is an efficient way to attenuate most ARGs in swine manure within the 6-day treatment, especially when compared with traditional composting. However, the gut environment also selects for increased horizontal gene transfer potential, which may make risk assessment of the use of vermicompost as a fertilizer more complex. As a novel biotechnology, vermicomposting with housefly larvae is still efficient at both reducing the volume of waste and the incidence of ARGs.
Accession codes
References
Alcock RE, Sweetman A, Jones KC . (1999). Assessment of organic contaminant fate in waste water treatment plants I: Selected compounds and physicochemical properties. Chemosphere 38: 2247–2262.
Aminov RI . (2011). Horizontal gene exchange in environmental microbiota. Front Microbiol 2: 158.
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA et al. (2008). The RAST server: Rapid annotations using subsystems technology. BMC Genomics 9: 75.
Bernard L, Chapuis-Lardy L, Razafimbelo T, Razafindrakoto M, Pablo AL, Legname E et al. (2012). Endogeic earthworms shape bacterial functional communities and affect organic matter mineralization in a tropical soil. ISME J 6: 213–222.
Binh CTT, Heuer H, Kaupenjohann M, Smalla K . (2009). Diverse aadA gene cassettes on class 1 integrons introduced into soil via spread manure. Res Microbiol 160: 427–433.
Blahovec J, Kostecka Z, Kocisova A . (2006). Peptidolytic enzymes in different larval stadium of housefly Musca domestica. Vet Med-Czech 51: 139–144.
Boucher Y, Labbate M, Koenig JE, Stokes HW . (2007). Integrons: mobilizable platforms that promote genetic diversity in bacteria. Trends Microbiol 15: 301–309.
Brinkmann N, Tebbe CC . (2007). Leaf-feeding larvae of Manduca sexta (Insecta, Lepidoptera) drastically reduce copy numbers of aadA antibiotic resistance genes from transplastomic tobacco but maintain intact aadA genes in their feces. Environ Biosafety Res 6: 121–133.
Calvert C, Morgan N, Martin R . (1970). House fly larvae: biodegradation of hen excreta to useful products. Poultry Sci 49: 588–589.
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336.
Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ et al. (2009). The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37: D141–D145.
Dominguez J, Aira M, Gomez-Brandon M . (2010). Vermicomposting: earthworms enhance the work of microbes. In: Insam H, Whittle IF, Goberna M (eds) Microbes at Work. Springer-Verlag: Berlin, Heidelberg, 93–114.
Egert M, Wagner B, Lemke T, Brune A, Friedrich MW . (2003). Microbial community structure in midgut and hindgut of the humus-feeding larva of Pachnoda ephippiata (Coleoptera: Scarabaeidae). Appl Environ Microbiol 69: 6659–6668.
Egert M, Marhan S, Wagner B, Scheu S, Friedrich MW . (2004). Molecular profiling of 16S rRNA genes reveals diet-related differences of microbial communities in soil, gut, and casts of Lumbricus terrestris L. (Oligochaeta: Lumbricidae). FEMS Microbiol Ecol 48: 187–197.
Enne VI, Livermore DM, Stephens P, Hall LMC . (2001). Persistence of sulphonamide resistance in Escherichia coli in the UK despite national prescribing restriction. Lancet 357: 1325–1328.
Eren AM, Maignien L, Sul WJ, Murphy LG, Grim SL, Morrison HG et al. (2013). Oligotyping: differentiating between closely related microbial taxa using 16S rRNA gene data. Methods Ecol Evol 4: 1111–1119.
Forsberg KJ, Reyes A, Bin W, Selleck EM, Sommer MOA, Dantas G . (2012). The shared antibiotic resistome of soil bacteria and human pathogens. Science 337: 1107–1111.
Gaze WH, Zhang LH, Abdouslam NA, Hawkey PM, Calvo-Bado L, Royle J et al. (2011). Impacts of anthropogenic activity on the ecology of class 1 integrons and integron-associated genes in the environment. ISME J 5: 1253–1261.
Ghosh S, LaPara TM . (2007). The effects of subtherapeutic antibiotic use in farm animals on the proliferation and persistence of antibiotic resistance among soil bacteria. ISME J 1: 191–203.
Hartenstein R, Hartenstein F . (1981). Physicochemical changes effected in activated sludge by the earthworm Eisenia foetida. J Environ Qual 10: 377–381.
Heck K, De Marco EG, Duarte MW, Salamoni SP, Van der Sand S . (2015). Pattern of multiresistant to antimicrobials and heavy metal tolerance in bacteria isolated from sewage sludge samples from a composting process at a recycling plant in southern Brazil. Environ Monit Assess 187: 328.
Hvistendahl M . (2012). Public health China takes aim at rampant antibiotic resistance. Science 336: 795–795.
Knights D, Kuczynski J, Charlson ES, Zaneveld J, Mozer MC, Collman RG et al. (2011). Bayesian community-wide culture-independent microbial source tracking. Nat Methods 8: 761–U107.
Langille MGI, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA et al. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31: 814–821.
Lemos FJA, Terra WR . (1991). Digestion of bacteria and the role of midgut lysozyme in some insect larvae. Comp Biochem Phys B 100: 265–268.
Li B, Yang Y, Ma LP, Ju F, Guo F, Tiedje JM et al. (2015a). Metagenomic and network analysis reveal wide distribution and co-occurrence of environmental antibiotic resistance genes. ISME J 9: 2490–2502.
Li DH, Liu CM, Luo RB, Sadakane K, Lam TW . (2015b). MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31: 1674–1676.
Li H, Durbin R . (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760.
Liu B, Pop M . (2009). ARDB-antibiotic resistance genes database. Nucleic Acids Res 37: D443–D447.
Liu QL, Tomberlin JK, Brady JA, Sanford MR, Yu ZN . (2008). Black soldier fly (Diptera: Stratiomyidae) larvae reduce Escherichia coli in dairy manure. Environ Entomol 37: 1525–1530.
Looft T, Johnson TA, Allen HK, Bayles DO, Alt DP, Stedtfeld RD et al. (2012). In-feed antibiotic effects on the swine intestinal microbiome. Proc Natl Acad Sci USA 109: 1691–1696.
Mayende L, Wilhelmi BS, Pletschke BI . (2006). Cellulases (CMCases) and polyphenol oxidases from thermophilic Bacillus spp. isolated from compost. Soil Biol Biochem 38: 2963–2966.
Mazel D . (2006). Integrons: agents of bacterial evolution. Nat Rev Microbiol 4: 608–620.
McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A et al. (2012). An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 6: 610–618.
Mendes LW, Kuramae EE, Navarrete AA, van Veen JA, Tsai SM . (2014). Taxonomical and functional microbial community selection in soybean rhizosphere. ISME J 8: 1577–1587.
Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M . (2007). KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res 35: W182–W185.
Mumcuoglu KY, Miller J, Mumcuoglu M, Friger M, Tarshis M . (2001). Destruction of bacteria in the digestive tract of the maggot of Lucilia sericata (Diptera: Calliphoridae). J Med Entomol 38: 161–166.
Naas T, Mikami Y, Imai T, Poirel L, Nordmann P . (2001). Characterization of In53, a class 1 plasmid- and composite transposon-located integron of Escherichia coli which carries an unusual array of gene cassettes. J Bacteriol 183: 235–249.
Neuhauser E, Kaplan D, Malecki M, Hartenstein R . (1980). Materials supporting weight gain by the earthworm Eisenia foetida in waste conversion systems. Agricul Wastes 2: 43–60.
Ozogul F . (2004). Production of biogenic amines by Morganella morganii, Klebsiella pneumoniae and Hafnia alvei using a rapid HPLC method. Eur Food Res Technol 219: 465–469.
Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW . (2015). CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25: 1043–1055.
Petridis M, Bagdasarian M, Waldor MK, Walker E . (2006). Horizontal transfer of shiga toxin and antibiotic resistance genes among Escherichia coli strains in house fly (Diptera: Muscidae) gut. J Med Entomol 43: 288–295.
Pfaffl MW . (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45.
Poole TL, Callaway TR, Bischoff KM, Warnes CE, Nisbet DJ . (2006). Macrolide inactivation gene cluster mphA-mrx-mphR adjacent to a class 1 integron in Aeromonas hydrophila isolated from a diarrhoeic pig in Oklahoma. J Antimicrob Chemoth 57: 31–38.
Ramsden S, Ghosh S, Bohl L, LaPara T . (2010). Phenotypic and genotypic analysis of bacteria isolated from three municipal wastewater treatment plants on tetracycline-amended and ciprofloxacin‐amended growth media. J Appl Microbiol 109: 1609–1618.
Rho MN, Tang HX, Ye YZ . (2010). FragGeneScan: predicting genes in short and error-prone reads. Nucleic Acids Res 38: e191.
Riber L, Poulsen PHB, Al-Soud WA, Hansen LBS, Bergmark L, Brejnrod A et al. (2014). Exploring the immediate and long-term impact on bacterial communities in soil amended with animal and urban organic waste fertilizers using pyrosequencing and screening for horizontal transfer of antibiotic resistance. FEMS Microbiol Ecol 90: 206–224.
Seemann T . (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30: 2068–2069.
Smillie CS, Smith MB, Friedman J, Cordero OX, David LA, Alm EJ . (2011). Ecology drives a global network of gene exchange connecting the human microbiome. Nature 480: 241–244.
Su JQ, Wei B, Xu CY, Qiao M, Zhu YG . (2014). Functional metagenomic characterization of antibiotic resistance genes in agricultural soils from China. Environ Interl 65: 9–15.
Su JQ, Wei B, Ou-Yang WY, Huang FY, Zhao Y, Xu HJ et al. (2015). Antibiotic resistome and its association with bacterial communities during sewage sludge composting. Environ Sci Technol 49: 7356–7363.
Su Z, Zhang M, Liu X, Tong L, Huang Y, Li G et al. (2010). Comparison of bacterial diversity in wheat bran and in the gut of larvae and newly emerged adult of Musca domestica (Diptera: Muscidae) by use of ethidium monoazide reveals bacterial colonization. J Economic Entomol 103: 1832–1841.
Wall DP, Deluca T . (2007). Ortholog detection using the reciprocal smallest distance algorithm. Methods Mol Biol 396: 95–110.
Wang FH, Ma WQ, Dou ZX, Ma L, Liu XL, Xu JX et al. (2006). The estimation of the production amount of animal manure and its environmental effect in China. China Environ Sci 26: 614–617.
Wang FH, Qiao M, Su JQ, Chen Z, Zhou X, Zhu YG . (2014). High throughput profiling of antibiotic resistance genes in urban park soils with reclaimed water irrigation. Environ Sci Technol 48: 9079–9085.
Wang H, Zhang Z, Czapar GF, Winkler MK, Zheng J . (2013). A full-scale house fly (Diptera: Muscidae) larvae bioconversion system for value-added swine manure reduction. Waste Manage Res 31: 223–231.
Warnecke F, Luginbuhl P, Ivanova N, Ghassemian M, Richardson TH, Stege JT et al. (2007). Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature 450: 560–U517.
Wood DE, Salzberg SL . (2014). Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol 15: R46.
Xia LC, Steele JA, Cram JA, Cardon ZG, Simmons SL, Vallino JJ et al. (2011). Extended local similarity analysis (eLSA) of microbial community and other time series data with replicates. BMC Syst Biol 5: S15.
Zhang Z, Wang H, Zhu J, Suneethi S, Zheng J . (2012). Swine manure vermicomposting via housefly larvae (Musca domestica): the dynamics of biochemical and microbial features. Bioresour Technol 118: 563–571.
Zhang ZJ, Shen JG, Wang H, Liu M, Wu LH, Ping F et al. (2014). Attenuation of veterinary antibiotics in full-scale vermicomposting of swine manure via the housefly larvae (Musca domestica). Sci Rep-Uk 4: 6844.
Zhu YG, Johnson TA, Su JQ, Qiao M, Guo GX, Stedtfeld RD et al. (2013). Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc Natl Acad Sci USA 110: 3435–3440.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (41373074, 21210008), Zhejiang Science and Technology Innovation Program (2013C33001, 2015C03SA420001), and the National Key Research and Development Plan (2016YFD0900205). This work was supported in part by the US Department of Energy under Contract DE-AC02-06CH11357.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Wang, H., Sangwan, N., Li, HY. et al. The antibiotic resistome of swine manure is significantly altered by association with the Musca domestica larvae gut microbiome. ISME J 11, 100–111 (2017). https://doi.org/10.1038/ismej.2016.103
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2016.103
This article is cited by
-
Effects of magnesium-modified biochar on antibiotic resistance genes and microbial communities in chicken manure composting
Environmental Science and Pollution Research (2023)
-
Prevalence of macrolide–lincosamide–streptogramin resistant lactic acid bacteria isolated from food samples
Journal of Food Science and Technology (2023)
-
Beyond canonical models: why a broader understanding of Diptera-microbiota interactions is essential for vector-borne disease control
Evolutionary Ecology (2023)
-
Can Antibiotic Resistance Genes in Household Food Waste be Reduced by Earthworm Vermicomposting? Underpinning Mechanisms and Strategies
Reviews of Environmental Contamination and Toxicology (2023)
-
Effects of Chinese medicine herbal residues on antibiotic resistance genes and the bacterial community in chicken manure composting
The Journal of Antibiotics (2022)