Abstract
Crops lack genetic resistance to most necrotrophic pathogens. To compensate for this disadvantage, plants recruit antagonistic members of the soil microbiome to defend their roots against pathogens and other pests. The best examples of this microbially based defense of roots are observed in disease-suppressive soils in which suppressiveness is induced by continuously growing crops that are susceptible to a pathogen, but the molecular basis of most is poorly understood. Here we report the microbial characterization of a Korean soil with specific suppressiveness to Fusarium wilt of strawberry. In this soil, an attack on strawberry roots by Fusarium oxysporum results in a response by microbial defenders, of which members of the Actinobacteria appear to have a key role. We also identify Streptomyces genes responsible for the ribosomal synthesis of a novel heat-stable antifungal thiopeptide antibiotic inhibitory to F. oxysporum and the antibiotic’s mode of action against fungal cell wall biosynthesis. Both classical- and community-oriented approaches were required to dissect this suppressive soil from the field to the molecular level, and the results highlight the role of natural antibiotics as weapons in the microbial warfare in the rhizosphere that is integral to plant health, vigor and development.
Similar content being viewed by others
Introduction
Plant disease-suppressive soils are the best examples of microbiologically based defense of roots against soil-borne pathogens. These are soils in which, because of their microbial makeup and activity, a pathogen does not establish or persist, establishes but causes little or no disease or establishes and causes disease at first but then the disease declines with successive cropping of a susceptible host crop (Cook and Rovira, 1976; Raaijmakers and Weller, 1988; Weller et al., 2002; Sanguin et al., 2009; Kwak and Weller, 2012). Suppressive soils have re-emerged as a major topic of research because of the growing emphasis on sustainable and organic agriculture, recognition of the need to reduce the use of chemical pesticides and the fact that they provide control of soil-borne pathogens with little or no off-farm inputs.
Classically, soil suppressiveness has been considered to be either general or specific, with the former owing to the collective competitive and antagonistic activity of the total soil microbiome and the latter owing to a specific group(s) of microorganisms acting against a specific pathogen and often on a specific crop. General suppression is not transferable, is reduced by soil steaming, is enhanced by practices that increase overall soil microbial activity and provides only a basal level of defense against a broad range of pathogens. In contrast, highly effective specific suppression is superimposed over general suppression, is transferable by adding as little as 1% of suppressive soil into conducive soil, is eliminated by soil pasteurization and acts against a specific pathogen and often on a specific crop. Long-term continuous monoculture of a crop can induce specific soil suppressiveness, and a break in monoculture will reduce or eliminate it, but suppression can rapidly be regained when the inducing crop is again grown (Weller et al., 2002; Weller, 2007). The microbiological and biochemical foundations of soil suppressiveness are a major focus of current research, in part because of new molecular tools that, in combination with more traditional approaches, have begun to shed light on interactions not only between suppressive microorganisms and target pathogens but also among members of entire rhizosphere microbial communities. It is expected that these approaches may provide insight into more complex arrays of microorganisms and mechanisms involved in the protective effects associated with specific suppressiveness than previously detected by conventional culture-based approaches alone.
Fusarium spp. have an extremely broad host range and are among the most devastating soil-borne pathogens in crop production systems worldwide, causing symptoms including damping-off, root rot and vascular wilt. Fusarium wilt of strawberry, caused by Fusarium oxysporum f. sp. fragariae, is particularly difficult to control, partly because the pathogen produces chlamydospores that persist in soil for years. In the current study, we use both classical and community-based approaches to dissect a Fusarium wilt-suppressive soil beginning in the field and ending at the cellular and molecular levels and show how one suppressive microbe disrupts the growth of fungal pathogens, leading to arrest of the disease.
Materials and methods
Determination of the incidence and severity of Fusarium wilt
To determine the suppressiveness of the soils, four treatments were compared: suppressive soil (S, 15-year strawberry monoculture soil); conducive soil (C, 3-year monoculture soil); mixed soil (CS, C:S=90%:10%); and pasteurized soil (SH, 15-year monoculture soil heat treated, 80 °C for 1 h). At the third planting (cycle), a fifth treatment with gamma-irradiated (60 KGy, KAERI, Daejon, Republic of Korea) 15-year monoculture soil was included (SR) as a microbe-free control. Inoculum of F. oxysporum chlamydospores was added to each soil at a dose of 2 × 105 cfu g−1 of soil. Soil was added to plastic pots (22 cm × 60 cm × 18 cm) and 50-day-old strawberry plants (cv. Janghee) were planted in each pot. Treatments were arranged in a randomized complete block design and replicated five times with a single pot serving as a replicate. Plants were incubated in a growth chamber (25 °C; 16 h light and 8 h dark cycle) and harvested 45 days after planting. The incidence and severity of wilt were evaluated on a 0–5 scale; 0=healthy, 1=1–3 leaves rolled and yellowed, 2=>3 leaves rolled and deformed, 3=chlorosis and early plant wilting, 4=necrosis and entire plant wilting, and 5=dead or nearly so. In addition, rhizosphere soil was collected by vigorously shaking the roots and then stored at −80 °C for later soil DNA extraction. The remainder of the soil from each pot was immediately returned to the pot, and a new seedling was planted. In total, there were three sequential plantings (cycles) of strawberry grown and evaluated in each pot.
Pyrosequencing, operational taxonomic unit (OTU) analysis and taxonomic assignment
Soil DNA was extracted from 1 g of rhizosphere soil from each of three plants of the 13 treatments in the cycling study using a FastDNA SPIN Kit (MP Biomedicals, Santa Ana, CA, USA) following the manufacture’s protocol. DNA for libraries was amplified from each replicate according to the GS-FLX plus library prep guide. Universal primers for 16 S (27 F (5′-GAGTTTGATCMTGGCTCAG-3′) and 518 R (5′-WTTACCGCGGCTGCTGG-3′)) were used to amplify bacterial 16S rRNA genes. The PCR products were purified using AMPure beads (Beckman Coulter, Brea, CA, USA), quantified using the Picogreen assay (Victor 3, PerkinElmer, Waltham, MA, USA), pooled and subjected to emulsion PCR by using the GS-FLX plus emPCR Kit (454 Life Sciences, Brandford, CT, USA) according to the manufacturer’s instructions. The immobilized libraries were suspended and emulsified in the emPCR product solution. The DNA-carrying beads were dispensed into a 96-well plate, and PCR amplification was performed. After emPCR, the libraries were purified using biotinylated primer A and the amplified single-stranded DNA library was quantified with a Particle Counter (Beckman Coulter). For pyrosequencing, the samples were loaded on a half-plate of Roche GX-FLX plus (Basel, Switzerland). Raw reads from each sample were passed through a data processing pipeline with the CD-HIT-OTU software (http://weizhong-lab.ucsd.edu/) to filter homopolymer error, chimera reads and denoising and then sorted by the tag barcode sequences. All the sequence data were aligned using the SILVA rRNA database (http://arb-silva.de). To find the optimum alignment, the Needleman–Wunsch global alignment algorithm was used. Mothur (www.mothur.org) and FitTree (Version 1.4) were used to visualize phylogenetic trees. Phylogenetic designations were generated with 75% similarity at the phylum level. Non-metric multi-dimensional scaling and the Bray–Curtis index were used to calculate community dissimilarity at the phylum level. Rhizomicrobiome sequence data were deposited in GenBank (accession number: SRP052821).
Genome analysis of Streptomyces sp. S4-7 and genome mining of secondary metabolites
The genome sequence of Streptomyces sp. S4-7 was determined by using the Illumina/Solexa platform (San Diego, CA, USA). A total of 5.2 and 4.4 Gb of sequences from a 363-bp paired-end library and a 8-kb mate-pair library were generated, respectively (Macrogen, Seoul, Republic of Korea). Sequence data were quality-trimmed and assembled with CLC Genomics Workbench version 4.8 (CLC Bio, Inc., Aarhus, Denmark). Construction of scaffolds and automatic gap closing were conducted by using the SSPCE version 1.2 (Boetzer et al., 2011; http://www.baseclear.com/genomics/bioinformatics/basetools/SSPACE) and IMAGE version 2.4 (Tsai et al., 2010; https://www.sanger.ac.uk). The final assembly led to 178 contigs with approximately 978-fold genome coverage. Gene prediction of the draft S4-7 genome sequence was conducted using Glimmer 3.0 (Delcher et al., 2007; https://ccb.jhu.edu/software/glimmer). Functional assignment of the predicted genes was achieved by searching for homologs in public protein databases, including UniRef100, Pfam, the KEGG, COG and GenBank non-redundant protein database, by using the BLAST program (Altschul et al., 1997). The outputs were automatically parsed using AutoFACT (Koski et al., 2005) and then complemented with the annotation results from the RAST server (Aziz et al., 2008). tRNA-scan (Lowe and Eddy, 1997) was applied to search for tRNA genes in the genome, and rRNA genes were identified by using BLAST. The sequence and annotation have been deposited in GenBank under the accession JYBE01000000. To obtain putative antibiotic information, the genome sequences were loaded into the Antibiotics and Secondary Metabolites Analysis Shell 2.0 (antiSMASH, http://antismash.secondarymetabolites.org) (Blin et al., 2013).
Molecular networking of Streptomyces sp. S4-7 secondary metabolites
The data files were converted to .mzXML format and subjected to the molecular networking workflow (http://gnps.ucsd.edu/ProteoSAFe/static/gnps-splash.jsp). Networks were constructed with a cosine threshold of 0.65 and a minimum of six matched peaks. The data were visualized with Cytoscape version 3.1.0; (http://www.cytoscape.org/).
Isolation and MS analysis of the thiopeptide
S4-7 was grown on PDK agar for 7 days, and the whole plate was cut into pieces and extracted with 10 ml n-butanol overnight with shaking (200 r.p.m.) at room temperature. The extract was filtered through a 0.2-μm membrane, dried, suspended in methanol and separated on a column of Sephadex LH20 (Pharmacia Fine Chemicals Inc., Piscataway, NJ, USA) eluted with methanol. The fractions were dried and suspended in 20 μl methanol. Mass spectrometry was carried out with high-resolution electrospray ionization tandem mass spectrometry (HR-ESI-MS/MS; Agilent Q-TOF (Agilent Technologies, Palo Alto, CA, USA), 0% acetonitrile/H2O (0.1% formic acid) to 100% acetonitrile, 25 min, 1000–3200 m/z, 20 keV) and Fourier Transform ion cyclotron resonance MS/MS direct injection (Advion BioSciences, Ithaca, NY, USA; 200–2000 m/z, 35 keV).
Chemogenomic studies of yeast heterozygous and homozygous genome-wide deletion pools
To determine the targets of the antifungal activity, wild-type yeast (BY4741) was used in growth-inhibition assays with cell-free S4-7 supernatant. To prescreen the supernatant, S4-7 was inoculated in PDK broth and incubated with shaking (200 r.p.m.) at 28 °C for 10 days. Cells were removed by centrifugation (10 000 r.p.m.; 10 min) and the supernatant was passed through a 0.22-μm filter (Santa Cruz Biotechnology, Santa Cruz, CA, USA). BY4741 cells were grown, diluted to an initial OD600 of 0.0625 and added to each well of a 24-well plate (VWR, Radnor, PA, USA). The cell-free S4-7 supernatant solution was added to each well to achieve concentrations of 0, 12.5, 25, 50 and 75% in a total volume of 1.4 ml. Plates were incubated for 24 h at 30 °C in a microplate spectrophotometer (BioTek, Winooski, VT, USA) with shaking, and every 15 min absorbance (OD600) was measured. For heterozygous and homozygous deletion profiling assays, the assays were performed in triplicate with different concentrations of cell-free S4-7 supernatant (12.5, 25 and 50%). The heterozygous and homozygous collections were grown for 20 and 5 generations, respectively. Cells were harvested by centrifugation and genomic DNA was extracted with DNeasy Blood and Tissue Kits (Qiagen, Valencia, CA, USA) according to the manufacturer’s protocol. PCR reactions to amplify barcode sequences from heterozygous or homozygous pooled genomic DNA used previously reported protocols (Smith et al., 2011). Briefly, the PCR mixture included 6 μl 10 × PCR buffer (without MgCl2), 3 μl 50 mM MgCl2, 1.2 μl 10 mM dNTPs, 1.2 μl 50 μM Up or Down primer mix, 0. 6 μl 5U μl−1 Taq polymerase and 0.1 μg genomic DNA in a total of 60 μl. The barcode sequences were amplified with the following conditions: initial denaturing at 94 °C for 3 min, 30 cycles of 94 °C for 30 s, 55 °C for 30 s, 72 °C for 30 s, and then 72 °C for 3 min as a final extension step. The PCR products were loaded on a gel and checked for amplicon size and quality. For hybridization, prewetted TAG4 arrays (Affymetrix, Santa Clara, CA, USA) were filled with 1 × hybridization buffer and incubated at 42 °C for 10 min at 20 r.p.m. A hybridization mixture was prepared as follows: 75 μl 2 × buffer, 0.5 μl B213 control oligonucleotide, 12 μl mixed oligonucleotides and 3 μl 50 × Denhardt’s solution to a volume of 90 μl. A total of 30 μl uptag PCR and 30 μl downtag PCR products was added to the hybridization mix, boiled for 2 min and set in ice-water for 2 min. The prehybridization buffer was removed and 90 μl hybridization mix with the PCR mixture was added. Hybridization was conducted at 42 °C for 16 h with shaking (20 r.p.m.). All washing steps of the arrays were performed with an Affymetrix fluidics station using the manufacturer’s instructions. The arrays were scanned with an emission wavelength of 560 nm in an Affymetrix GeneArray scanner. Outlier masking, removing unusable tags, saturation correction, array normalization and calculation sensitivity scores for control–treatment comparisons were performed as previously reported (Hillenmeyer et al., 2008). For manual individual validation, the deletion strains with log2 ratio of ⩾5 from the deletion collection assays were selected. The selected strains were mixed with cell-free S4-7 supernatant (0, 12.5 25 and 50%, total volume 1.4 ml) in each well of 24-well plates. The strains were incubated, and growth was monitored as described above. The validation experiments were conducted with four replicates. To find bioactive compounds with similar mode-of-action, the fitSearch algorithm (Lee et al., 2013) was used (http://fitsearch.kaist.ac.kr). Gene Ontology (GO) slim analysis and GO full annotations were performed according to the SGD website (http://www.yeastgenome.org). Physical–genetic interactions of the direct target protein candidates were analyzed in GeneMANIA (http://www.genemania.org).
Results
Verification of a Fusarium wilt-suppressive soil
We investigated two commercial strawberry fields located in Sancheong County, Korea and observed that a 15-year continuous monoculture field showed a significantly lower incidence of Fusarium wilt than did a 3-year monoculture field located only a few meters away (Supplementary Figures S1a and b). The two sites differed somewhat in the contents of sand, silt and clay. The 3-year monoculture field soil contained more total carbon and nitrogen than did the 15-year monoculture soil. In contrast, the 15-year monoculture field soil had more Cu and Zn than did soil from the 3-year monoculture field (Supplementary Table S1). Disease symptoms in the 3-year strawberry field included irregular leaf development, destruction of vascular tissue, crown rot and stunting as compared with healthy plants (Figures 1a–d). The observation of side-by-side fields differing markedly in disease severity is typical of how specific suppressive soils historically have been identified. In traditional pot studies, soils from the two fields also differed strikingly in their specific suppressiveness against F. oxysporum: wilt was suppressed when strawberry was planted into the 15-year soil but not in the 3-year soil. To demonstrate the transferability of the specific suppressiveness, the 3- and 15-year soils were mixed in a ratio of 9:1 (9C:1 S), respectively. The mixture showed substantially less suppression of wilt after one cycle than did the 15-year soil alone (Figure 1e), but in the second and third successive plantings (cycles) into the same pots, the 9C:1 S mixture showed the same level of suppressiveness as the 15-year soil. In all three cycles, strawberry in the 3-year soil had severe disease, and there was no evidence of disease suppression (Figures 1e–g). Pasteurized 15-year soil (80 °C, 1 h) lost wilt suppressiveness after one cycle of strawberry, but suppression returned to the level of the 15-year soil in the second and third successive plantings (cycles) into the same pots (Figures 1e–g), indicating that the heat treatment reduced but did not completely eliminate the suppressive agents. Gamma-irradiation (Figure 1g), however, completely eliminated suppressiveness. Collectively, these results are consistent with microbially based specific suppressiveness resulting from a relatively slow-growing, heat-tolerant but irradiation-sensitive agent(s).
Fusarium wilt of strawberry and evaluation of specific suppressiveness. Typical symptoms of Fusarium wilt: (a) stunting, (b) crown rot, (c) deformed leaves as compared with (d) a healthy plant. Successive crops (e) first, (f) second and (g) third of strawberry were planted into monoculture soils, and chlamydospores of F. oxysporum were inoculated at a dose of 2 × 105 cfu g−1 of soil. Wilt disease was evaluated 45 days later. S, 15-year strawberry monoculture soil; C, 3-year monoculture soil; CS, soil mix (3- and 15-year monoculture soils mixed 9:1 w/w); S80, heat-treated 15-year soil (80 °C, 1 h); SR, gamma-irradiated 15-year soil. Bars in the same panel with the same letter are not significantly different (LSD; P<0.05).
Rhizobacterial communities and isolation of a representative suppressive agent
To begin to identify the agent(s) involved, we used next-generation sequencing to compare the microbial community structure in each strawberry rhizosphere. From the 13 cycled samples, we obtained 54 000–79 000 reads/sample passing quality-control filters (Supplementary Table S2), with a total of 842 039 reads and an average read length of 427 base pairs. Rarefaction curves indicated that the number of reads was sufficient to assess the diversity of organisms in the rhizosphere communities (Supplementary Figure S2a), and taxonomic analysis identified 17 taxa at the phylum level (Supplementary Figure S2b). Among the 13 rhizosphere samples, 13 084 OTUs were detected by using a 3% dissimilarity cutoff. The most abundant phyla included Proteobacteria (40.31%), followed by Firmicutes (13.6%), Actinobacteria (11.36%) and Acidobacteria (10.4%). These four phyla made up >63% of rhizomicrobiome, with unclassified phyla accounting for an additional 3.41% of detected OTUs (Supplementary Figure S2b).
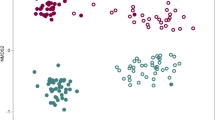
The diversity and abundance of phyla in the 13 rhizomicrobiomes, as examined by Shannon and Simpson indices, revealed no significant differences in bacterial community composition (Supplementary Table S3), but non-metric multi-dimensional analysis indicated that the soils could be separated according to their disease suppressiveness in successive cycling experiments (Figure 2a), suggesting that the microbial community differed functionally among the various samples. No one phylum was selectively and consistently enriched by cycling or in the heat-treated soils (Supplementary Table S4–S6), nor did a relative abundance matrix show a perfect match with the pattern of disease suppressiveness (data not shown). Among the top 15 rhizobacterial OTUs more abundant in all the suppressive soils than in the conducive soils were Actinobacteria (six OTUs), Proteobacteria (six OTUs), Acidobacteria (two OTUs), Gemmatimonadetes (one OTU) and Nitrospirae (one OTU) (Table 1). Among the top 84 OTUs shared in all the suppressive soils and more abundant than in the conducive soils, Actinobacteria accounted for 26% followed by Acidobacteria (22%), Proteobacteria (20%), Chloroflexi (14%) and Gemmatimonadetes (11%), with the remaining 7% belonging to unknown or other phyla (Supplementary Figure S3).
Microbial community analysis and root colonization by Streptomyces sp. S4-7. (a) Non-metric multi-dimensional scaling ordination of rhizosphere microorganisms. The matrix was generated using the Bray–Curtis similarity algorithm. Numbers indicate each planting cycle, and S refers to 15-year suppressisve soil, C represents 3-year conducive soil, CS indicates mixed soil; heat-treated 15-year soil is shown as S80, and SR indicates gamma-irradiated 15-year soil. Filled symbols indicate disease-suppressive soils and the disease-conducive soils are represented by open symbols. (b) Population density of Streptomyces sp. S4-7 in the rhizosphere. S4-7 was suspended in 0.1% methyl cellulose (MC) solution to a density of 3.5 × 107 cfu ml−1. S: Streptomyces sp. S4-7 (pIJ10257); F: chlamydospores (2 × 105 cfu g−1 of soil) of F. oxysporum; SF: F. oxysporum and Streptomyces sp. S4-7 (pIJ10257). The roots were dipped for 2 min in each treatment before planting. (c) Disease severity was evaluated on a 0–5 scale. Root colonization and disease severity were evaluated 3, 5 and 7 weeks after inoculation.
Because the results of the cycling experiments suggested that at least one agent important in wilt disease suppression had unique characteristics, including slow growth and tolerance to treatment at 80 °C, and considering that among the OTUs more abundant in the suppressive than in the conducive soils (Table 1), only the Actinobacteria met these criteria, we next used selective media to recover these bacteria from the rhizosphere of strawberry grown in the 15-year soil and then evaluated the isolates in vitro for antagonism against F. oxysporum. Strain S4-7, selected as representative of these Actinobacteria, showed >95% 16S sequence similarity to Streptomyces sp. SXY07 (GU045526), a strain included among the OTUs more abundant in suppressive soils than in conducive soils (Table 1).
Disease suppressiveness and rhizosphere colonization by Streptomyces sp. S4-7
In growth chamber and commercial field studies, introduced Streptomyces sp. S4-7 showed an exceptional ability to suppress Fusarium wilt (Supplementary Figures S1c and d). We tracked the population dynamics of S4-7 in the rhizosphere by introducing the plasmid pIJ10257 to generate a hygromycin-resistant derivative strain. This recombinant showed the same level of antifungal activity as the wild type and could be followed by using selective media followed by PCR amplification to detect the introduced selective marker. The recombinant microbe maintained a population density above 105 cfu g−1 of root for up to 7 weeks regardless of whether or not the pathogen was present (Figure 2b). Rhizosphere colonization is critical for disease suppression, and 105 cfu g−1 of root is the threshold density required for disease suppression (Raaijmakers and Weller, 1988). Similar to the wild-type strain, the recombinant provided robust protection of strawberry against wilt (Figure 2c).
Genome mining of Streptomyces sp. S4-7 and identification of a novel thiopeptide involved in suppressiveness
Based on sequencing data, the Streptomyces sp. S4-7 genome is 7.6 Mb in size with 71.57% G+C content (Supplementary Figure S4a). BLAST comparisons showed that the genome is most similar and syntenic to that of Streptomyces griseus NBBR3350, with an average nucleotide identity value of 90.39% as estimated by JSpecies version 1.2.1 (Richter and Rosselló-Móra, 2009; Supplementary Figure S4b). Heat-denatured (70 °C, 5–15 min) Streptomyces sp. S4-7 supernatant fully retained the ability to inhibit yeast growth (Supplementary Figure S4c), indicating the synthesis of a thermostable and diffusible metabolic product. Therefore, we analyzed the genome of S4-7 using antiSMASH (Blin et al., 2013) and identified 35 gene clusters encoding putative antimicrobial natural products representing a variety of structural classes (Supplementary Table S7).
To determine which of these clusters might produce metabolite(s) responsible for disease suppressiveness, we mutated S4-7 and isolated two classes of mutants in which antifungal activity was completely abolished. The first mutation identified a 34-kb gene cluster (Figure 3a) predicted to produce a novel ribosomally synthesized thiopeptide. This chemically diverse family is known to contain members with potent antimicrobial activity (Yu et al., 2009; Arnison et al., 2013; Wang et al., 2013). Bioinformatic analysis of this thiopeptide cluster revealed a putative 58 amino-acid precursor peptide with a C-terminus rich in threonine, serine and cysteine residues (Supplementary Table S8) as well as two peptidases not previously known to be involved in thiopeptide maturation. As the thiopepetide had not previously been reported, we assigned the name conprimycin, meaning suppressive in Latin.
Novel thiopeptide antibiotic responsible for pathogen inhibition. (a) Thiopeptide mutants (ΔT-1, ΔT-2, ΔT-3) lost antagonistic activity against F. oxysporum. Bioinformatic analysis of the partial thiopeptide gene cluster (contig 93). ▴ indicates the position of a knockout in ORF 8. (b) Loss of antifungal activity in an ectoine mutant and structure of the ectoine biosynthesis gene cluster in contig 22. ▴ shows the knockout position in ORF 6. (c) Structure of the partially dehydrated and cyclized thiopeptide. Fragments detected by mass spectrometry are highlighted in red. (d) Alignment of the proposed S4-7 thiopeptide precursor with characterized thiopeptide precursors. (e) Disease-suppressive assay with wild-type S4-7 and mutants. Macrospores of F. oxysporum (1 × 107 cfu g−1 of soil) were inoculated and disease severity was evaluated on a 0–5 scale.
A second mutation was identified in a cluster predicted to produce ectoine (Figure 3b), a pyrimidine carboxylic acid derivative that functions as a compatible solute and protein chaperone in many bacteria (Pastor et al., 2010). Inactivation of genes in the thiopeptide and ectoine loci (Supplementary Figures S4d and e) confirmed that the loss of antifungal activity (Figures 3a and b) was due to the loss of products from these clusters. Mutants in both loci completely lacked antifungal activity in vitro and failed to fully protect plants against pathogen attack in growth chamber assays (Figure 3e and Supplementary Figure S5).
To identify the product of the conprimycin gene cluster, cell extracts were prepared from wild-type Streptomyces sp.S4-7 as well as the conprimycin and ectoine mutant strains and analyzed by high-performance liquid chromatography–HR-ESI-MS/MS. The metabolomic information, analyzed by molecular networking (Frank et al., 2008; Watrous et al., 2012; Yang et al., 2013), revealed a doubly charged ion at m/z 1107.053 in the wild type. The same peak was absent not only from the conprimycin mutant but also from the ectoine mutant (Supplementary Figure S6a). To further connect the compound to its gene cluster, a peptidogenomic (Kersten et al., 2011) approach was used to reveal peptide fragments consistent with the postulated biosynthetic product (Figures 3c and d and Supplementary Figure S6b).
A chemogenomic approach reveals the mode of action of S4-7
A key facet of antagonist/pathogen interactions that has escaped characterization in work on suppressive soils is the mode of action by which a microbial agent inhibits its target pathogen. To address this question, we interrogated the yeast Saccharomyces cerevisiae deletion collection against S4-7 cell-free supernatants, an approach that bypasses the need for pure antifungal compound(s) produced by the antagonist. We observed a dose-dependent response against wild-type yeast (BY4741) using S4-7 extracts (Supplementary Figure S7a). For global chemogenomic profiling, we performed a HIP-HOP (HaploInsufficiency Profiling-Homozygous deletion Profiling) assay (Giaever et al., 1999) with increasing concentrations of S4-7 extracts. Following analysis of the heterozygous deletion pool (typically used to uncover putative targets via drug-induced haploinsufficiency) (Giaever et al., 1999; Lopez et al., 2008), we selected 22 mutant yeast strains (HIP) based on their degree of sensitivity and confirmed the growth inhibition by individual strain analysis (log2 ratio>5, Figure 4a and Supplementary Table S9). GO slim analysis (slim terms with >2 proteins, Supplementary Figure S7b) showed that the proteins encoded by the deletion strains were primarily involved in cell wall organization and biogenesis (PKC1, SBE2, HSP150, CDA2), transcription (SOH1, YGR122W, GCN5, ARP9), cytoskeleton organization and RNA catabolism. To further focus our list of putative targets, we posited that growth inhibition was likely to be dependent on supernatant concentration as well as target protein amount (Supplementary Table S10). This approach revealed that two cell wall synthesis proteins, PKC1 and SBE2, were most likely to be directly affected by S4-7 extracts (Figure 4b; Supplementary Figure S7b). Interestingly, both proteins are functionally linked by several physical and genetic interactions and are networked with other gene products, including SOH1, GCN5 via HSP150 and cytoskeleton organization (RVS161, AIM3) via CDA2 (Figure 4b). To find bioactive compounds with a similar mode of action, we compared the profile of a homozygous yeast deletion pool (HOP) treated with S4-7 supernatant to profiles generated by bioactive compounds in the FitSearch database (Lee et al., 2013). Bleomycin and rapamycin, natural products of Streptomyces and the marine sponge compounds latrunculin and papuamide B (Supplementary Table S11), have a mode-of-action profile similar to that of S4-7.
Chemogenomic studies with heterozygous deletion mutants of yeast and biological interaction map. (a) Growth curves of wild-type yeast strain BY4741 and selected deletion strains in the presence of cell-free supernatant of S4-7. Growth inhibition of other strains is described in Supplementary Table S10. Open symbols indicate yeast grown without supernatant of S4-7, and closed symbols indicate treatment with S4-7 supernatant. (b) Physical (in red) and genetic (in green) interactions between yeast proteins assigned to GO slim terms. Line thickness is proportional to confidence level provided by GeneMANIA (http://www.genemania.org).
Discussion
Plant and microbe species diversity in natural ecosystems have a critical role in maintaining low levels of disease (Gilbert, 2002), but crop rotation, tillage, and the cultivation of limited numbers of crop varieties in modern farming favor the incidence and severity of diseases caused by necrotrophic soil-borne pathogens. With successive planting of the same crop, however, soils can achieve a state known as specific suppressiveness due to the selection and enrichment over time of particular microbial populations that are transferrable to nonsuppressive soils and typically are eliminated by pasteurization, fumigation or crop rotation. In our study, the 15-year strawberry monoculture soil exhibited transferability after more than one planting cycle, retained activity after heating at 80 °C and required irradiation to fully eliminate control of Fusarium wilt, suggesting the involvement of heat-resistant and comparatively slow-growing microorganisms in the soil’s suppressiveness. These results are consistent with molecular evidence revealing that Actinobacteria were more abundant in suppressive than in the conducive soils (Table 1). Additionally, strain S4-7 colonized the roots of strawberry well, as would be expected if it had been selected by long-term monoculture. Collectively, our classical approach, combining soil cycling to transfer slower-growing protective populations, soil irradiation to inactivate heat-resistant strains and microbial community analysis to reveal the presence over time of Actinobacteria in the rhizomicrobiome, enabled us to target these bacteria as candidates with an important role in this novel example of specific soil suppressiveness and culminated in the discovery of a representative suppressive strain, Streptomyces S4-7.
As others have found in community-based analyses of suppressive soils (Sanguin et al., 2009; Mendes et al., 2011; Rosenzweig et al., 2012), no single phylum was uniquely associated with disease suppression in the strawberry wilt microbiome (Table 1 and Supplementary Tables S4, S5 and S6). Instead, our identification of antibiotic-producing Streptomyces as participating in the suppression of Fusarium wilt also required culture-based approaches that have long implicated specific microorganisms in those instances where the etiology of specific soil suppressiveness has been at least partially resolved. Thus, whereas fluorescent Pseudomonas spp. producing the antibiotic 2,4-diacetylphloroglucinol have a key role in the suppression of take-all disease of wheat known as take-all decline in US soils and soils in the Netherlands (Weller et al., 2002; Weller, 2007) and nonpathogenic Streptomyces spp. have long been known to contribute to the suppression of common scab of potato caused by pathogenic Streptomyces spp. (Bowers et al., 1996; Lorang et al., 1989), community-based studies of such soils have shown that multiple taxa are associated with suppression but have not resolved functionally active agents at the species or subspecies level in either pathosystem (Sanguin et al., 2009; Rosenzweig et al., 2012). A third study, guided by a community-based approach and on which our work was modeled, also identified multiple phyla in a soil suppressive of damping-off disease caused by Rhizoctonia solani, but culture-based methods were needed to identify specific strains of an antibiotic-producing Pseudomonas inhibitory to the pathogen (Mendes et al., 2011). Until now, community-based approaches have revealed both a greater degree of microbial complexity in suppressive soils than previously was recognized and that the structure and association networks of taxa can shift in parallel with microbial community functional characteristics (Bakker et al., 2014). However, the identification of functionally relevant phyla is still constrained by limitations in databases and the resolving power of the present technology, and the taxa that currently are quantifiable are not necessarily the most functionally relevant ones. These limitations, and the many still-unresolved questions about physical and biochemical interactions in the rhizosphere, await new tools to unravel ecological basis of the below-ground relationships that govern soil suppressiveness. Hartmann et al. (2015) also have recently commented on the difficulty of inferring the ecological role of many microbial community members simply from phylogenetically based surveys.
Given their abundance in soil and the diversity of broad-spectrum antibiotics they produce, it is surprising that actinomycetes have received comparatively little attention as players in rhizosphere interactions with plant pathogens. Prominent exceptions are the Streptomyces species involved in the specific suppressiveness of field soils to common scab of potato after long-term potato cultivation (reviewed by Weller et al., 2002) and Streptomyces griseoviridis, a major component of the commercial biofungicide MYCOSTOP (Lahdenperä et al., 1991). The latter strain produces a candicidin polyene macrolide antibiotic, genes for the synthesis of which are widely distributed among the Actinobacteria. Thiopeptide biosynthesis operons also are wide spread in Actinobacteria, suggesting that these antibiotics have an important ecological role in nature. Another such thiopeptide compound, cyclothiazomycin B1, has been reported to have heat-stable antibiotic properties and to control fungal plant pathogens via disruption of cell wall chitin molecules (Mizuhara et al., 2011) and inhibition of RNA polymerase (Hasimoto et al., 2006). The size of the novel thiopeptide identified in our study, as well as fragmentation analysis of fractions in which it was present, revealed several amino-acid mass shifts corresponding to the sequence of the predicted thiopeptide and support the connection of the compound to the thiopeptide gene cluster. It also is tempting to speculate that the mature S4-7 thiopeptide contains several macrocyclic ring systems consistent with other model thiopeptides such as thiostrepton and nosiheptide, as there are no further linear fragmentation patterns visible from MS. The newly discovered compound is unusually large for a thiopeptide and appears to be the largest member of this group reported to date.
Our data indicate that the bioactive compound produced by S4-7 acts to block fungal cell wall biogenesis and stability, resulting in growth inhibition. The target PKC1 is a protein serine/threonine kinase essential for cell wall remodeling during growth, and SBE2 is also involved in the transport of cell wall components from the Golgi to the cell surface. Our physical and genetic interaction map shows that these targets are functionally linked to each other as well as to transcription (SOH1, GCN5) via HSP150 (a heat shock protein covalently attached to the cell wall and required for cell wall stability) and cytoskeletal organization (RVS161, AIM3) via CDA2 (chitin deacetylase, required for rigidity of the ascospore wall). FitSearch (Lee et al., 2013) profiles of latrunculin, papuamide B, belomycin and rapamycin are similar to that of the S4-7 thiopeptide, and their antifungal mechanisms of action (Supplementary Table S12) appear to be tightly linked to cellular functions of the target candidates of S4-7, including the cell wall/membrane (papuamide and bleomycin), cytoskeleton organization (latrunculin) and cell growth and protein synthesis through a signaling pathway (rapamycin).
In summary, we have characterized a 15-year continuous monoculture strawberry field soil that is highly suppressive to Fusarium wilt, demonstrating that our observations are consistent with Actinobacteria producing a thiopeptide associated with suppression and showing that a major target of the antibiotic is cell wall synthesis. Our study defines a framework for characterizing components of the soil microbiome that protect roots against attack by fungal pathogens. The application of broadened classical methods traditionally used to study suppressive soils, along with modern community analysis and bioanalytical techniques, allowed a full and rapid dissection of the workings of this wilt-suppressive soil from the field to the cellular level. Suppressive soils occur globally for all classes of soil-borne pathogens and provide a highly sustainable means to control pests using minimal off-farm inputs. Such soils are ideal model systems in which to unravel the ‘black box’ of in situ plant–microbe interactions and the microbial warfare in the rhizosphere that are integral to plant health, vigor and development. Given that plants routinely lose >21% of all photosynthate from their roots and soil-borne pests reduce crop yields by ⩾5–60% annually, elucidating the plant–microbe interactions in the rhizosphere required for optimal root and soil health will be critical to launching the next ‘Green Revolution’ needed to feed the world’s population that may double by the end of the twenty-first century.
Accession codes
References
Altschul SF, Maddern TL, Schäffer AA, Zhang J, Zhang Z, Willer W et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucl Acids Res 25: 3389–3402.
Arnison PG, Bibb MJ, Bierbaum G, Bowers AA, Bugni TS, Bulaj G et al. (2013). Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat Prod Rep 30: 108–160.
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA et al. (2008). The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9: 75.
Bakker MG, Schlatter DS, Otto-Hanson L, Kinkel LL . (2014). Diffuse symbioses: roles of plant-plant, plant-microbe, and microbe-microbe interactions in structuring the soil microbiome. Mol Ecol 23: 1571–1583.
Blin K, Medema MH, Kazempour D, Fischbach MA, Breitling R, Takano E et al. (2013). antiSMASH 2.0-a versatile platform for genome mining of secondary metabolite producers. Nucl Acids Res 41: W204–W212.
Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W . (2011). Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 27: 578–579.
Bowers J, Kinkel L, Jones R . (1996). Influence of disease-suppressive strains of Streptomyces on the native Streptomyces community in soil as determined by the analysis of cellular fatty acids. Can J Microbiol 42: 27–37.
Cook RJ, Rovira AD . (1976). The role of bacteria in the biological control of Gaeumannomyces graminis by suppressive soils. Soil Biol Biochem 8: 269–273.
Delcher AL, Bratke KA, Powers EC, Salzberg SL . (2007). Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23: 673–679.
Frank AM, Banderia N, Shen Z, Tanner S, Briggs SP, Smith RD et al. (2008). Clustering millions of tandem mass spectra. J Proteome Res 7: 113–122.
Giaever G, Shoemaker DD, Jones TW, Liang H, Winzeler EA, Astromoff A et al. (1999). Genomic profiling of drug sensitivities via induced haploinsufficiency. Nat Genet 21: 278–283.
Gilbert GS . (2002). Evolutionary ecology of plant diseases in natural ecosystem. Ann Rev Phytopathol 40: 13–43.
Hartmann M, Frey B, Mayer M, Mäder P, Widmer F . (2015). Distinct soil microbial diversity under long-term organic and conventional farming. ISME J 9: 1177–1194.
Hasimoto M, Murakami T, Funahashi K, Tokunaga T, Nihei K, Okuno T et al. (2006). An RNA polymerase inhibitor, cyclothiazomycin B1, and its isomer. Bioorg Med Chem 14: 8259–8270.
Hillenmeyer ME, Fung E, Wildenhain J, Pierce SE, Hoon S, Lee W et al. (2008). The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science 320: 362–365.
Kersten RD, Yang YL, Xu Y, Cimermancic P, Nam SJ, Fenical W et al. (2011). A mass spectrometry-guided genome mining approach for natural product peptidogenomics. Nat Chem Biol 7: 794–802.
Koski LB, Gray MW, Lang BF, Burger G . (2005). AutoFACT: an automatic functional annotation and classification tool. BMC Bioinformatics 6: 151.
Kwak YS, Weller DM . (2012). Take-all of wheat and natural disease suppression: a review. Plant Pathol J 29: 125–135.
Lahdenperä M-L, Simon E, Uoti J . (1991). Mycostop® - a novel biofungicide based on Streptomyces bacteria. In: Beemster ABR, Bollen GJ, Gerlagh M, Ruissen MA, Schippers B, Tempel A (eds). Biotic Interactions and Soil-Borne Diseases. Elsevier: : Amsterdam, The Netherlands, pp 258–263.
Lee M, Han S, Chang H, Kwak YS, Weller DM, Kim D . (2013). FitSearch: a robust way to interpret a yeast fitness profile in terms of drug's mode-of-action. BMC Genomics 14: S6.
Lopez A, Parsons AB, Nislow C, Giaever G, Boone C . (2008). Chemical-genetic approaches for exploring the mode of action of natural products. Prog Drug Res 66: 239–271.
Lorang J, Anderson N, Lauer F, Wildung D . (1989). Disease decline in a Minnesota potato scab plot. Am Potato J 66: 531.
Lowe TM, Eddy SR . (1997). tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucl Acids Res 25: 955–964.
Mendes R, Kruijt M, de Brujin I, Dekkers E, vand der Voort M, Schneider JH et al. (2011). Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332: 1097–1100.
Mizuhara N, Kuroda M, Ogita A, Tanaka T, Usuki Y, Fujuta K . (2011). Antifungal thiopeptide cyclothiazomycin B1 exhibits growth inhibition accompanying morphological changes via binding to fungal cell wall chitin. Bioorg Med Chem 19: 5300–5310.
Pastor JM, Salvador M, Argandoña M, Bernal V, Reina-Bueno M, Csonka LN et al. (2010). Ectoines in cell stress protection: Uses and biotechnology production. Biotechnol Adv 28: 782–801.
Raaijmakers JM, Weller DM . (1988). Natural plant protection by 2,4-diacetylphloroglucinol-producing Pseudomonas spp. in take-all decline soils. Mol Plant Microbe Interact 11: 144–152.
Richter M, Rosselló-Móra R . (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA 106: 19126–19131.
Rosenzweig N, Tiedje JM, Quensen JE III, Meng Q, Hao JJ . (2012). Microbial communities associated with scab-suppressive soils determined by pyrosequencing analyses. Plant Dis 96: 718–725.
Sanguin H, Sarniguet A, Gazengel K, Moënne-Loccoz Y, Grundmann GL . (2009). Rhizosphere bacterial communities associated with disease suppressive stages of take-all decline in wheat monoculture. New Phytol 184: 694–707.
Smith AM, Durbic T, Oh J, Urbanus M, Proctor M, Heisler LE et al. (2011). Competitive genomic screens of barcoded yeast libraries. J Vis Exp 54: e2864.
Tsai IJ, Otto TD, Berriman M . (2010). Improving draft assemblies by iterative mapping and assembly of short reads to eliminate gaps. Genome Biol 11: R41.
Wang S, Zhou S, Liu W . (2013). Opportunities and challenges from current investigations into the biosynthetic logic of nosiheptide-represented thiopeptide antibiotics. Curr Opin Chem Biol 17: 626–634.
Watrous J, Roach P, Alexandrov T, Heath BS, Yang JY, Kersten RD et al. (2012). Mass spectral molecular networking of living microbial colonies. Proc Natl Acad Sci USA 109: E1743–E1752.
Weller DM . (1988). Biological control of soilborne plant pathogens in the rhizosphere with bacteria. Annu Rev Phytopathol 26: 37–407.
Weller DM . (2007). Pseudomonas biocontrol agents of soilborne pathogen: looking back over 30 years. Phytopathology 97: 250–256.
Weller DM, Raaijmakers JM, McSpadden Gardener BB, Thomashow LS . (2002). Microbial populations responsible for specific soil suppressiveness to plant pathogens. Annu Rev Phytopathol 40: 309–348.
Yang JY, Sanchez LM, Rath CM, Liu X, Boudreau PD, Bruns N et al. (2013). Molecular networking as a dereplication strategy. J Nat Prod 76: 1686–1699.
Yu Y, Duan L, Zhang Q, Liao R, Ding Y, Pan H et al. (2009). Nosiheptide biosynthesis featuring a unique indole side ring formation on the characteristic thiopeptide framework. ACS Chem Biol 4: 855–864.
Acknowledgements
Part of this work was supported by RDA of Korea (PJ010827). Other funding included Royal Society, UK (516002.K5677/ROG) to HH, an NRF grant (NRF-2011-0017670) to JFK, NIH grant (GM97509) to BSM, BK21 PLUS to HC and S-KK and a DFG postdoctoral fellowship to MC. We thank Tim Paulitz for helpful discussions.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Cha, JY., Han, S., Hong, HJ. et al. Microbial and biochemical basis of a Fusarium wilt-suppressive soil. ISME J 10, 119–129 (2016). https://doi.org/10.1038/ismej.2015.95
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2015.95
This article is cited by
-
Characterization of rhizospheric fungi and their in vitro antagonistic potential against myco-phytopathogens invading Macrotyloma uniflorum plants
International Microbiology (2024)
-
Intercropping wheat alleviated soil acidification and suppressed Fusarium wilt of faba bean
Plant and Soil (2024)
-
Repeated exposure of wheat to the fungal root pathogen Bipolaris sorokiniana modulates rhizosphere microbiome assembly and disease suppressiveness
Environmental Microbiome (2023)
-
Acidification suppresses the natural capacity of soil microbiome to fight pathogenic Fusarium infections
Nature Communications (2023)
-
Identifying microbiota community patterns important for plant protection using synthetic communities and machine learning
Nature Communications (2023)