Abstract
Marine sediments are the largest carbon sink on earth. Nearly half of dark carbon fixation in the oceans occurs in coastal sediments, but the microorganisms responsible are largely unknown. By integrating the 16S rRNA approach, single-cell genomics, metagenomics and transcriptomics with 14C-carbon assimilation experiments, we show that uncultured Gammaproteobacteria account for 70–86% of dark carbon fixation in coastal sediments. First, we surveyed the bacterial 16S rRNA gene diversity of 13 tidal and sublittoral sediments across Europe and Australia to identify ubiquitous core groups of Gammaproteobacteria mainly affiliating with sulfur-oxidizing bacteria. These also accounted for a substantial fraction of the microbial community in anoxic, 490-cm-deep subsurface sediments. We then quantified dark carbon fixation by scintillography of specific microbial populations extracted and flow-sorted from sediments that were short-term incubated with 14C-bicarbonate. We identified three distinct gammaproteobacterial clades covering diversity ranges on family to order level (the Acidiferrobacter, JTB255 and SSr clades) that made up >50% of dark carbon fixation in a tidal sediment. Consistent with these activity measurements, environmental transcripts of sulfur oxidation and carbon fixation genes mainly affiliated with those of sulfur-oxidizing Gammaproteobacteria. The co-localization of key genes of sulfur and hydrogen oxidation pathways and their expression in genomes of uncultured Gammaproteobacteria illustrates an unknown metabolic plasticity for sulfur oxidizers in marine sediments. Given their global distribution and high abundance, we propose that a stable assemblage of metabolically flexible Gammaproteobacteria drives important parts of marine carbon and sulfur cycles.
Similar content being viewed by others
Introduction
Marine coastal sediments are global hot spots of carbon remineralization and burial (Hedges and Keil, 1995). In current models of oceanic carbon cycling, the sequestration of microbially altered organic matter is the major mechanism of carbon preservation in sediments (Parkes et al., 1993; Burdige, 2007). Marine sediments are sites not only of carbon remineralization but also of carbon fixation. Recent estimates suggest that marine microbes fix inorganic carbon independent of light (chemolithoautotrophy) in amounts that are in the same order of magnitude as the annual organic carbon burial (Middelburg, 2011). Chemolithoautotrophic microorganisms in marine sediments fix up to 370 Tg C/year, which equals 48% of carbon fixed chemolithoautotrophically in the ocean (Middelburg, 2011). Thereof, 47% are fixed in shallow, near-shore sediments (175 Tg C/year). Near-shore sediments, therefore, contribute more to oceanic carbon fixation than pelagic oxygen minimum zones (OMZs) and hydrothermal vents (Middelburg, 2011). In recent years, chemolithoautotrophy in these marine systems has received much attention. The ecophysiology and genetic composition of key players of carbon (and sulfur) cycling in OMZs and hydrothermal vents, such as the gammaproteobacterial SUP05 clade, have been extensively studied (Lavik et al., 2009; Canfield et al., 2010; Reinthaler et al., 2010; Swan et al., 2011; Grote et al., 2012; Anantharaman et al., 2013; Mattes et al., 2013). This cosmopolitan clade is expected to have an important role in attenuating atmospheric carbon dioxide concentrations, when OMZs expand in a warming climate (Hawley et al., 2014).
In contrast to pelagic OMZs, where the oxic-anoxic/sulfidic interface can be meters thick, in near-shore sediments this interface is only a few millimeters thick, and is characterized by steep biogeochemical gradients and approximately 1000-fold higher cell abundances per sample volume. Biogeochemical evidence indicates that sulfur oxidation is the dominant chemolithoautotrophic process in coastal sediments, while nitrification appears to play only a minor role (Middelburg, 2011; Boschker et al., 2014). Previous studies of benthic autotrophic sulfur oxidizers mostly focused on large, conspicuous sulfur bacteria such as Beggiatoa, which are widely distributed but occur in high abundances only in certain habitats (Salman et al., 2013; Ruff et al., 2015). Other Gammaproteobacteria affiliating with cultured sulfur oxidizers (Acidithiobacillus, Thiohalophilus and Thiomicrospira) or with uncultured symbiotic sulfur oxidizers among the Chromaticaeae and Ectothiorhodospiracea have been regularly found in marine and estuarine sediments (Bowman et al., 2005; Orcutt et al., 2011). Consistent with this, recent molecular and isotopic approaches suggest that some of these are indeed autotrophs (Lenk et al., 2011; Boschker et al., 2014; Vasquez-Cardenas et al., 2015).
Culture-independent molecular studies previously identified predominant carbon fixation pathways such as the Calvin-Benson-Bassham (CBB) cycle and the reductive tricarboxylic acid cycle in marine chemolithoautotrophic bacteria. The key genes encoding ribulose-1,5-bisphosphate carboxylase-oxygenase (RuBisCO) form I and form II (cbbL, cbbM) and the ATP citrate lyase (aclAB) in the reductive tricarboxylic acid cycle pathway have been frequently detected in environmental studies (reviewed by Hügler and Sievert, 2011). Likewise, genes encoding subunits of the reverse dissimilatory sulfite reductase (dsrAB), of the adenosine-5'-phosphosulfate reductase (aprA) and of the thiosulfate-oxidizing Sox-multienzyme complex (soxB) have been used to target the diversity of marine benthic sulfur oxidizers (Lenk et al., 2011; Thomas et al., 2014).
To understand how inorganic carbon at sediment surfaces is turned over and possibly buried, detailed knowledge of the microbes driving these processes is essential but currently still lacking. To fill this gap, we surveyed the diversity of candidate bacterial chemolithoautotrophs in 13 coastal surface sediments across Western Europe and Australia. Moreover, we studied whether these chemolithoautotrophic bacteria are also present in anoxic, 490-cm-deep subsurface sediments. We developed a new method to combine 14C-bicarbonate labeling of cells with fluorescence in situ hybridization (FISH), fluorescence-activated cell sorting (FACS) and scintillography to quantify dark carbon fixation by distinct taxonomic groups. Meta- and single-cell genomics along with metatranscriptomics provided evidence for a largely sulfur-based carbon fixation in a selected tidal sediment. Metatranscriptomic reads were mapped against reference databases containing cbbL, cbbM and aclAB sequences to identify active carbon fixation pathways. Metatranscriptomic reads mapped against reference databases containing dsrAB, aprA and soxB sequences indicated sulfur oxidation pathways active in situ. This unique combination of molecular and isotopic approaches provided unprecedented insights into the ecology and ecophysiology of cosmopolitan microorganisms driving a major part of global dark carbon fixation.
Materials and methods
Sediment sampling and characteristics
Between October 2012 and December 2014, we sampled 10 tidal and 3 sublittoral sandy sediments in Western Europe and Australia (Figure 1 and Supplementary Table S1). The 10 tidal sediments were sampled during low tide using polyacryl-cores or cutoff syringes of up to 25 cm length. The three sublittoral, coastal sandy sediments were sampled during cruise He417 with the RV Heincke in March 2014 in the German Bight using multi- and boxcorers. At each site, two to three different sediment layers were selected for molecular analyses (16S rRNA gene amplicon sequencing, catalyzed reporter deposition (CARD)-FISH). Sediment for 14C incubations was collected from sites Calais, Courseulles-sur-Mer and from Janssand (Supplementary Table S1). Sediment colors served as a proxy for redox state and active sulfide formation and oxidation (Figure 1). For most of the sites, sediment (i) from the uppermost cm (brownish sediment, sulfide-free), (ii) from the sulfide transition zone (brown to gray, reflecting the presence of iron sulfides) and (iii) from sediment of the sulfidic layer was sampled for molecular analyses (Supplementary Table S1). Sulfide concentrations in pore waters were measured for sites Calais and Courseulles-sur-Mer using the methylene blue method (Cline, 1969). More details on the biogeochemistry of sulfide and oxygen at the study sites Janssand and Königshafen in the German Wadden Sea have been published previously (de Beer et al., 2005; Billerbeck et al., 2006; Jansen et al., 2009). In addition, in April 2005 we sampled a 490-cm-deep subsurface core at site Janssand as described in detail by Gittel et al. (2008). Sulfate and methane concentration profiles and lithological data from this core have been described previously (Gittel et al., 2008; Seidel et al., 2012).
Sampling sites of the 16S rRNA gene survey (a). Example for a typical stratification of a sediment core from coastal sandy sediments (b). A=uppermost sediment layer, B=sulfide transition zone and C=sulfidic layer refer to the different sampling depths in this study. During sampling, the sediment colors were used as an indicator for the presence of iron sulfide (dark gray to black). Asterisk indicates samples from sublittoral sediments.
DNA extraction for barcoded 16S rRNA gene amplicon sequencing
For all intertidal and subsurface sediment samples from Europe and Australia, DNA was extracted from 200 to 250 μl sediment recovered from distinct layers using the PowerSoil DNA isolation kit (MoBio Laboratories, Solana Beach, CA, USA). DNA from sites NoahA, NoahB and CCPδ was extracted from 5 g of homogenized surface sediments according to Zhou et al. (1996) including Proteinase K treatment for improved cell lysis.
Barcoded 16S rRNA gene amplicon sequencing
The bacterial diversity in all sediment samples was determined by analyzing the hypervariable V3–V4 region of the 16S rRNA gene using Roche 454 pyro- or Illumina MiSeq-sequencing of barcoded amplicons. Barcoded amplicons from all surface sediments were prepared using primers 341f/785rev (Herlemann et al., 2011; Klindworth et al., 2012). Barcoded amplicons from the 490 cm-deep subsurface sediment from site Janssand were prepared by replacing primer 785rev with 907rev (Muyzer et al., 1998; Klindworth et al., 2012). This primer covers a similar bacterial diversity as the reverse primer 785rev used for surface sediments (see above, Figure 2), but it is known to bias against some phyla that are, however, low abundant or absent in marine sediments (Klindworth et al., 2012). In total, 311 196 bacterial 16S rRNA reads were kept for taxonomic classification using the SILVAngs pipeline v115 (Quast et al., 2013) with a clustering at 98% identity. Details on PCR conditions, sequencing and processing are given as Supplementary Information.
Relative abundances of the bacterial 16S rRNA gene sequence (V3–V4 regions) in the 13 coastal sediments. Left panels: relative sequence abundance of most frequent bacterial phyla and classes. Right panels: relative sequence abundance of most frequent taxonomic groups (family to order level) within the class of Gammaproteobacteria according to the taxonomy of SILVA release 117 (Pruesse et al., 2007). The five candidate chemolithoautotrophic clades are given in bold. Upper panels: relative sequence abundance in 13 coastal surface sediments from 0 to 23 cm below surface (cmbsf) max. A=uppermost sediment layer (0.5 or 1 cmbsf), B=sulfide transition zone, C=sulfidic layer. For the detailed depth ranges, see Supplementary Table S1. Lower panels: relative sequence abundance in subsurface sediments at site Janssand (JS) from 50 to 490 cmbsf. For comparison, samples from 10 cmbsf are included. Sediment samples from 10 and 490 cmbsf were run in duplicate (10ab, 490ab). All but one amplicons were pyrosequenced, samples from DB were sequenced via the Illumina MiSeq platform.
Sediment incubations with 14C-bicarbonate
Sediment for 14C-DIC incubations was collected from sites Calais and Courseulles-sur-Mer in July 2013 and from Janssand in April 2014 (Supplementary Table S1). These cores were kept at in situ temperature (15–20 °C) and used for 14C-DIC incubations within 48 h after sampling. After slicing, 2 ml of the uppermost sediment layer and from the sulfide transition zone was transferred into 10 ml glass vials. In all, 1 ml of sterile filtered seawater and 1 ml of artificial seawater containing 1.5 mm 14C bicarbonate (specific activity 54.7 mCi mmol−1, Hartmann Analytic, Braunschweig, Germany) were added. Vials were sealed with lab-grade butyl rubber stoppers (GMT Inc., Ochelata, OK, USA) leaving 6 ml of headspace (air). Then, vials were incubated for 20 h with mild agitation (100 r.p.m.) at in situ temperature in the dark. In parallel incubations 1 mm thiosulfate was added to slurries from Courseulles-sur-Mer and Calais. Slurry incubations were performed in duplicates (Calais, Courseulles-sur-Mer) or in triplicates (Janssand). Dead controls were included for each site by adding formaldehyde (2%, final concentration) before the incubation.
CARD-FISH and sample preparation for flow cytometry
For CARD-FISH, sediment from sites Calais, Courseulles-sur-Mer and Janssand was fixed immediately after core retrieval as described in Lenk et al. (2011). Cells were detached from 100 to 200 μl sediment by ultrasonic treatment as described previously (Lenk et al., 2011). Permeabilization and CARD-FISH were performed as described by Pernthaler et al. (2002) with the modifications detailed in Supplementary Information. Tyramides labeled with Alexa488 fluorescent dye (Molecular Probes, Eugene, OR, USA) were used for CARD signal amplification. An overview of oligonucleotide probes used in this study is shown in Supplementary Table S2. Novel oligonucleotide probes were designed for the JTB255 group using ARB and the SILVA 16S rRNA reference database release 117 (Pruesse et al., 2007). Please note that the Xanthomonadales, which includes the JTB255 clade, are not targeted by probe GAM42a, specific for most Gammaproteobacteria (Siyambalapitiya and Blackall, 2005). In line with this, the JTB255 clade could not be detected with probe GAM42a in double hybridizations with the JTB-probe mix (Supplementary Table S2). Therefore, we summed up FISH counts of the JTB255-probe mix and of probe GAM42a to yield the total relative abundance of FISH-detectable Gammaproteobacteria.
FACS and scintillography of sorted cells
We developed a novel protocol to quantify bulk assimilation of radiolabelled substrates in a defined number of cells phylogenetically identified via CARD-FISH before for flow sorting. To minimize cell loss, the filters were handled extremely carefully during the CARD-FISH procedure. Cells were scraped off from membrane filters by using a cell scraper or membrane filters were vortexed in 5 ml of 150 mm NaCl containing 0.05% Tween-80 according to Sekar et al. (2004). Before flow cytometry, large suspended particles were removed by filtration through 8-μm pore-size filter (Sartorius, Göttingen, Germany) to avoid clogging of the flow cytometer.
Flow sorting of cells that were fluorescently labeled by CARD-FISH was performed using a FACSCalibur flow cytometer equipped with cell sorter and a 15-mW argon ion laser exciting at 488 nm (Becton Dickinson, Oxford, UK). Autoclaved Milli-Q water was used as sheath fluid. Cell sorting was done at low flow rate of 12±3 μl min−1 or med flow rate of 35±5 μl min−1 with single-cell sort mode to obtain the highest purity. The event rate was adjusted with a fluorescence threshold, and sorting was performed at a rate of approximately 25–100 particles s−1. Hybridized cells were identified on scatter dot plots of green fluorescence versus 90° light scatter (Supplementary Figure S1). Sediment background such as clay particles was determined by flow-cytometric analysis of sediment hybridized with a nonsense probe (NON338) (Supplementary Figure S1). For subsequent measurements, 50 000 cells were sorted and filtered onto 0.2-μm polycarbonate filters (GTTP, Millipore, Eschborn, Germany). Dead controls of 14C incubated sediments did not show any measurable assimilation of 14C, and were thus not used for cell sorting. Unspecifically adsorbed label in live samples caused only minor radioactive background as determined by spiking experiments with fluorescent beads and Escherichia coli cells (Supplementary Figure S2). Fluorescent beads (yellow-green, 1.0 μm, Polyscience, Warrington, PA, USA) or hybridized E. coli cells were flow-sorted from of a sample hybridized with the nonsense probe to determine radioactive background. E. coli cells were hybridized with EUBI-III probe beforehand. Beads and E. coli cells were mixed with the sample in approximately the same quantity as the target populations (10–20% of total cells). Two to three repeated sortings were applied to confirm the technical reproducibility from duplicate or triplicate incubations. Collected cell batches on polycarbonate filters were directly transferred into 5 ml scintillation vials and mixed with 5 ml UltimaGold XR (Perkin-Elmer, Boston, MA, USA) scintillation cocktail. Radioactivity of sorted cell batches was measured in a liquid scintillation counter (Tri-Carb 2900, Perkin-Elmer).
The purity of flow-cytometric enriched target cells was >93%, and was manually analyzed under an Axioplan epifluorescence microscope (Zeiss, Jena, Germany). For microscopic analysis, filters were counterstained with 1 μg ml−1 4′,6-diamidino-2-phenylindole (DAPI) and at least 1000 DAPI-stained cells were examined for CARD-FISH. Our approach was technically highly reproducible, and the radioactivity linearly increased with the number of sorted cells (Supplementary Figure S2).
For calculation of average cell-specific carbon fixation rates in our slurry experiments, we assumed a background concentration of dissolved inorganic carbon of 2 mm (Billerbeck et al., 2006) as we used local seawater for our experiment (see Supplementary Information for calculations). The relative abundance of assimilating gammaproteobacterial cells in the sulfide transition zone from Calais, Courseulles-sur-Mer and Janssand sediments was determined by microautoradiography (MAR). MAR was performed according to Alonso and Pernthaler (2005) and Lenk et al. (2011) with an exposure time of 2 days. Relative abundance of MAR-positive cells was manually determined under an Axioplan epifluorescence microscope (Zeiss).
A single-cell genome of the SSr clade from Janssand sediment
In January 2011, the upper two centimetres of Janssand sediment were sampled for extraction and sorting of single bacterial cells for whole-genome amplification. After extraction, cells were cryopreserved with N,N,N-trimethylglycine (‘glycine betaine’) (Sigma-Aldrich, St Louis, MO, USA) at a final concentration of 4% according to Cleland et al. (2004), stored at −80 °C and shipped overseas. Single-cell sorting and whole-genome amplification via multiple displacement amplification were performed at the Bigelow Laboratory Single Cell Genomics Center (https://scgc.bigelow.org) as described by Swan et al. (2011). A single amplified genome (SAG) encoding a single, high quality 16S rRNA gene sequence affiliating with the SSr clade was sent to Max Planck Genome Centre (MP-GC) Cologne for MiSeq (Illumina) sequencing yielding 9 557 547 PE reads. The SAG assemblies were auto-annotated using the Joint Genome Institute IMG-ER pipeline (Markowitz et al., 2012). Details on cell extraction, sequencing and quality control of the assembled genomic data are given as Supplementary information.
cDNA libraries and metatranscriptomic mapping
In April 2013, sediment was sampled from the sulfide transition zone at site Janssand and immediately frozen on dry ice. Total RNA was extracted from sediment in triplicates (one ml each) by the Vertis Biotechnologie AG (Freising, Germany), and bacterial rRNA was depleted with the Ribo-Zero Magnetic Kit (for Bacteria) (Epicentre, Madison, WI, USA). Barcoded RNA TrueSEQ libraries were constructed from RNA extractions and paired-end sequenced using Illumina HiSeq2000 (MP-GC, Cologne, Germany). After quality trimming at a Phred score 28 using Nesoni clip v.0.115 (http://www.vicbioinformatics.com/software.nesoni.shtml), reads were mapped to reference databases of nucleotide sequences encoding key genes for sulfur oxidation (Sox-multienzyme complex, soxB; reverse dissimilatory sulfite reductase, dsrAB; adenosine-5′-phosphosulfate reductase, aprA; uptake [NiFe]-hydrogenase, hupL; ammonia monooxygenase, amoA, ribulose-1,5-bisphosphate carboxylase/oxygenase form I, cbbl, and form II, cbbm; ATP citrate lyase, aclAB) and to the SAG using Bowtie2 (Langmead and Salzberg, 2012). Details on program settings and normalization are given as Supplementary information.
Nucleotide accession numbers
All nucleotide sequences obtained in this study have been deposited in GenBank. Sequences of 16S rRNA and hydrogenase gene libraries are available under accession numbers KR824952–KR825244 and KR534775–KR534844, respectively. Amplicon sequences from the 16S rRNA gene surveys were deposited in NCBI BioProjects PRJNA283163 and PRJNA285206. All cDNA reads are available in BioProject PRJNA283210. Fosmid end sequences are available in NCBI's Genome Survey Sequences database (GSS) with the accession numbers KS297884–KS307053. The genome sequence of the SAG WSgam209 is accessible under the IMG Genome ID 2609459745 through the Joint Genome Institute portal IMG/ER (https://img.jgi.doe.gov/cgi-bin/er/main.cgi), the metagenomic bin Acidiferrobacter-a7 is accessible under the IMG Genome ID 2616644801.
Results and Discussion
Identification of candidate chemolithoautotrophs in coastal surface sediments
To identify candidate chemolithoautotrophs in coastal sediments, we studied the bacterial diversity in 10 tidal and 3 sublittoral sandy sediments from Western Europe and Australia (Figure 1 and Supplementary Table S1). We sequenced the V3–V4 region (>300 bp) of tagged 16S rRNA gene amplicons. After quality trimming, 311 196 Illumina- and 454-tag reads were recovered. Taxonomic classification revealed that Gammaproteobacteria were consistently among the most abundant clades on class to phylum level, accounting for 12–45% of sequences regardless of sampling site, sediment depth or season (Figure 2). These data were supported by CARD-FISH, which showed that Gammaproteobacteria make up 19–22% of all bacteria at sites Janssand, Calais and Courseulles-sur-Mer (Supplementary Table S5). At all sites, we observed a recurring diversity pattern also at the family to order level within the Gammaproteobacteria. We consistently identified candidate chemoautotrophs most closely related to: (1) Acidiferrobacter thiooxydans of the family Ectothiorhodospiraceae, (2) symbionts of the siboglinid tubeworms such as Oligobrachia spp. (henceforth designated as Siboglinidae Symbionts related, SSr, see Supplementary Figures S3 and S4), (3) ciliate symbiont Candidatus Thiobios zoothamnicoli and (4) BD7-8 clade, including the γ3 symbiont of the marine gutless oligochaete Olavius algarvensis (Woyke et al., 2006). Sulfur-dependent chemolithoautotrophy for cultured or symbiotic relatives of these clades has been shown before (Rinke et al., 2006; Lösekann et al., 2008; Hallberg et al., 2011; Kleiner et al., 2012). Acidiferrobacter thiooxydans can also grow autotrophically with ferrous iron (Hallberg et al., 2011). Moreover, we previously showed carbon fixation by the SSr- and the Acidiferrobacter-related clades, and determined relative cell abundances of up to 8% at site Janssand in the German Wadden Sea (Lenk et al., 2011).
Strikingly, in all tested sediments up to 52% of gammaproteobacterial sequences grouped with the uncultured JTB255 clade. This clade is affiliated with the order Xanthomonadales and accounted for the largest fraction of gammaproteobacterial sequences at 10 out of 13 sites (Figure 2). In line with the sequence data, CARD-FISH targeting members of the JTB255 clade revealed rod-shaped cells (Supplementary Figure S5) that made up 3–6% of total cell counts in Janssand, Calais and Courseulles-sur-Mer sediments (Supplementary Table S6). So far, the exact environmental function of the JTB255 clade is unknown;however, a sulfur-oxidizing activity has been hypothesized (Bowman and McCuaig, 2003). In summary, we identified five candidate chemolithoautotrophs that accounted for 28–75% of Gammaproteobacteria and for 8–31% (average=17%) of all bacterial sequences across all sites. Other potentially autotrophic populations such as sulfur-oxidizing Epsilonproteobacteria, anoxygenic phototrophs, the BD1-5/SN-2 clade, nitrifiers and cyanobacteria were found in low abundance or were patchily distributed (Figure 2).
Gammaproteobacteria in subsurface sediments
Because of the high sedimentation rates of >3 mm/year in the German Wadden Sea at site Janssand (Ziehe, 2009), a yet unknown fraction of the surface microbial community including the chemolithoautotrophic Gammaproteobacteria is buried into the anoxic subsurface. To study, how these organisms are affected by such a burial, we also analyzed the distribution of chemolithoautotrophic Gammaproteobacteria in a 490-cm-deep subsurface core from site Janssand, spanning a sedimentation record of 1000–2000 years (Ziehe, 2009). This sediment core displayed a typical sulfate-methane-transition zone in 150–200 cm below surface (cmbsf) (Gittel et al., 2008), reflecting the changes in the major metabolic pathways that are active along this depth range. Surprisingly, Gammaproteobacteria including the chemolithoautotrophic gammaproteobacterial clades also accounted for a dominant fraction of 16S rRNA gene pyrotags over the entire depth range (21–37% of all sequences, Figure 2). This observation was supported by 16S rRNA gene libraries from 200 and 490 cmbsf (Supplementary Figure S4), in which these clades accounted for 95 out of 289 clones (33%). To gain PCR-independent support for the dominance of Gammaproteobacteria in subsurface sediments, we fosmid-cloned large metagenomic fragments of ~40 kb in size from 490 cmbsf and taxonomically classified the end sequences (Supplementary Table S7), as FISH is commonly too insensitive to comprehensively target subsurface organisms with very low ribosome content (Schippers et al., 2005). In support of our 16S rRNA gene data, 24% of all prokaryote-affiliated fosmid end sequences (n=4052) showed best hits to Gammaproteobacteria, while only approximately 1% affiliated with Archaea (Supplementary Table S7).
Collectively, these molecular data indicate a relatively stable community structure over the 5-m depth range, despite the measured strong biogeochemical gradients. These data are consistent with similar total cell abundances and a similar composition of major phospholipids over the entire 490 cm depth in the same sediment core (Gittel et al., 2008; Seidel et al., 2012).
Whether these gammaproteobacterial populations are active despite very different biogeochemical settings in the subsurface, or whether they are simply surviving in subsurface sediments with little or no turnover, are currently unclear. Upon burial, marine microbial cells may survive in the subsurface over geological time scales without significant growth (Jørgensen, 2011). Energy for maintenance and survival could be supplied by fermentation of refractory organic matter and by the slow transport and diffusion of dissolved organic compounds to subsurface sediments, which has been demonstrated for the subsurface at site Janssand (Røy et al., 2008; Seidel et al., 2012).
Global occurrence of candidate chemolithoautotrophic Gammaproteobacteria
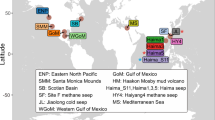
Our 16S rRNA diversity and FISH data agree well with numerous studies showing a substantial contribution of Gammaproteobacteria to microbial communities in diverse marine surface sediments (Hunter et al., 2006; Kim et al., 2008; Schauer et al., 2009; Orcutt et al., 2011; Gobet et al., 2012; Ruff et al., 2015). To examine the geographic distribution of the five candidate chemoautotroph groups in more detail, we did a meta-analysis of 16S rRNA gene sequence data from 65 diversity studies of the sea floor (Figure 3). Although these published data sets hardly covered the extent of microbial diversity at the studied sites, sequences related to the Acidiferrobacter, SSr and to a lesser extent Ca. T. zoothamnicoli and BD7-8 groups were found in all types of benthic habitats ranging from intertidal sediments to deep-sea hydrothermal chimneys (Figure 3). Intriguingly, the JTB255 clade was detected in 92% of all studies (Figure 3) and accounted for the most frequent sequence group among Bacteria in Arctic, Antarctic and tropical deep-sea as well as shallow coastal sediments (Wang et al., 2013; Zheng et al., 2014; Liu et al., 2014; Emil Ruff et al., 2014). In temperate Tasmanian and in cold Antarctic coastal sediments, 16S rRNA gene copies of the JTB255 clade accounted for 6–9% of total bacterial 16S rRNA gene sequences (Bowman et al., 2005). In summary, our biogeographic survey shows that these five clades are important members of microbial communities in marine surface sediments worldwide, and clearly, the JTB255 clade is one of the most successful bacterial lineages in marine surface sediments.
Biogeographic survey of the major chemolithoautotrophic gammaproteobacterial clades identified in this study. Only clone sequences (presence/absence) from bacterial diversity studies from 65 marine sea floor surfaces have been considered, deposited in the SILVA database release 117 (Pruesse et al., 2007).
Measuring dark carbon fixation by Gammaproteobacteria in sediments
Our 16S rRNA gene survey suggested that sulfur-oxidizing Gammaproteobacteria are potentially the major carbon fixers in dark coastal sediments. However, quantitative data on carbon fixation by distinct bacterial populations in marine sediments are lacking. To determine the contribution of Gammaproteobacteria to dark carbon fixation in sediments, we developed a novel approach to quantify assimilation of a radiolabeled compound by specific populations. Previous FACS experiments with autofluorescent, radiolabeled marine bacterioplankton and subsequent scintillography of sorted populations prompted us to use FISH signals instead of autofluorescence or unspecific DNA staining to identify and enrich populations from sediments (Zubkov et al., 2003; Jost et al., 2008). We incubated aerobic sediment slurries prepared from surface and from sulfide transition zone sediments from sites Calais, Courseulles-sur-Mer and Janssand with 14C-labeled bicarbonate and for 20 h in the dark. After detachment of cells from sand grains and CARD-FISH, we sorted fluorescently labeled Bacteria (probe EUBI-III) or Gammaproteobacteria (probe GAM42a). To account for potentially nitrifying autotrophic Archaea, we also sorted cells targeted by the archaeal probe Arch915. Fifty thousand cells were sorted per population, and bulk radioactivity was measured. This workflow allowed us to accurately quantify the bulk assimilation of radiolabeled substrates by a defined population in high throughput. At the analysis level of populations and communities it thereby overcomes limitations in throughput and precision of other methods such as MAR-FISH, HISH-SIMS and stable isotope probing (Boschker et al., 1998; Lee et al., 1999; Radajewski et al., 2000; Manefield et al., 2002; Musat et al., 2008).
Although the total amount of fixed carbon in sorted populations varied between sites and samples, the 14C-assimilation by sorted Gammaproteobacteria ranged from 2.4 to 10.3 Bq and was 2.5- to 5-fold higher than those of sorted Bacteria (0.5–2.1 Bq) (Figure 4a). At all three sites, the relative abundance of 14C-assimilating Gammaproteobacteria was approximately 40–50% as determined by MAR (Supplementary Table S8; Lenk et al., 2011) and is similar to the relative sequence frequency of chemoautotrophic subpopulations (Figure 2). The 14C-radioactivity of 50 000 archaeal cells accounted for 1.4–3.8 Bq, ranging between the average assimilation by Bacteria and Gammaproteobacteria (Figure 4b). Addition of nitrate did not stimulate 14C-assimilation in anoxic slurry incubations. The addition of 1 mm thiosulfate doubled the total carbon fixation by Gammaproteobacteria in the uppermost surface sediments, but not in the sulfide transition zone at Calais and Courseulles-sur-Mer (Figure 4a). Likewise, thiosulfate did not stimulate carbon fixation in the sulfide transition zone from site Janssand (Zerjatke, 2009). The oxidized surface sediments are possibly limited in electron donors, while the sulfide transition zone contained sufficient reduced sulfur compounds such as free and iron sulfides as energy sources for carbon fixation (Jansen et al., 2009; Supplementary Table S1).
14C carbon fixed by Bacteria, Gammaproteobacteria and Archaea in three coastal sediments. Carbon fixation by flow-sorted populations of Bacteria , Gammaproteobacteria and Archaea from the uppermost sediment layer (surface, 0–1 cmbsf) and from the sulfide transition zone (trans.) incubated with 14C bicarbonate. 14C-assimilation by Bacteria and Gammaproteobacteria at three sampling sites (Courseulles-sur-Mer, Calais and Janssand) and in different seasons (a). 14C-assimilation by three gammaproteobacterial clades and Archaea (b). Batches of 50 000 cells were sorted per measurement. The 14C carbon activity is given in Becquerel (Bq).
Dark carbon fixation in Gammaproteobacterial clades
To quantify carbon fixed by the candidate chemolithoautotrophic clades, we used the FISH probes available for three of the five clades for cell sorting and subsequent scintillography. Our FISH probes for the SSr and Acidiferrobacter clades mostly target sequences retrieved from site Janssand (Supplementary Figure S4; Lenk et al., 2011); therefore, we measured carbon assimilation by specific subpopulations at this site. The Acidiferrobacter clade showed the highest 14C-assimilation (6.1–10 Bq), while the SSr clade assimilated 2.7–6.9 Bq (Figure 4b and Supplementary Figure S6). This is consistent with the chemolithoautotrophic potential encoded in the corresponding genomes (Supplementary Table S5), and is confirmed by the previously detected 14C bicarbonate assimilation by single cells of both clades (Lenk et al., 2011).
Our probes for the ubiquitous JTB255 clade display a wider target range; therefore, we used these probes to sort JTB255 cells from Janssand, Calais and Courseulles-sur-Mer. The 14C-assimilation by the JTB255 clade ranged from 3 to 4.4 Bq in Janssand sediment to up to 10.7 Bq in Calais sediment (Figure 4b and Supplementary Figure S6). The 14C-assimilation by the JTB255 clade was slightly less than that of the SSr clade. The addition of thiosulfate did not stimulate the 14C-assimilation by the JTB255 clade in Courseulles-sur-Mer but did slightly stimulate it in Calais sediments, which is consistent with the hypothesized thiotrophy of members of this clade (Bowman and McCuaig, 2003).
The 14C-assimilation by the three gammaproteobacterial clades was up to 10-fold higher than the 14C-assimilation by the entire bacterial community (Figures 4a and b), which largely consisted of heterotrophic bacteria (Figure 2). Using the carbon assimilated by the bulk bacterial community (targeted by probes EUBI-III) as an approximate reference for heterotrophic, anaplerotic carbon fixation (Wood and Werkman, 1936; Li, 1982; Roslev et al., 2004), we conclude that heterotrophic carbon fixation was minor. Moreover, we calculated average carbon fixation rates per sorted gammaproteobacterial cell based on all sorted Gammaproteobacteria (1.1–3.0 fg C cell−1 day−1, Supplementary Table S8). Average carbon fixation rates per cell and for each of the three individual subpopulations ranged from 1.1 to 3.5 fg C cell−1 day−1. These rates are consistent with those of autotrophic freshwater green sulfur bacteria (1.4–5.8 fg C cell−1 day−1) (Musat et al., 2008), and are in the lower range of rates measured for autotrophic marine bacterioplankton (3.5–24.7 fg C cell−1 day−1) (Jost et al., 2008). Collectively, these data strongly support an autotrophic carbon fixation by the Acidiferrobacter, SSr and JTB255 clades.
Gammaproteobacteria dominate dark carbon fixation in coastal sediments
The relative contribution of carbon fixed by the Acidiferrobacter, SSr and JTB255 clades amounted to 77% of gammaproteobacterial and to 50–62% of bacterial dark carbon fixation at Janssand (Figure 5). Although they make up only 19–22% of the total microbial community, Gammaproteobacteria in total accounted for 70–86% of the microbial dark carbon fixation irrespective of sampling site, season and sediment depth (Figure 5 and Supplementary Figure S7).
Relative contribution of Gammaproteobacteria to total bacterial dark carbon fixation in three coastal sediments. The relative contribution of carbon fixed by total Gammaproteobacteria and by the Acidiferrobacter, SSr and JTB255 clade to total bacterial dark carbon fixation was calculated by integrating the 14C-carbon assimilation per population (dark gray bar) and the relative cell abundances of the respective populations in the sediments. Error bars represent the standard deviation (s.d.) of 3–4 replicate incubations. Values from 2009 (site Janssand) are depicted as average of duplicates only. Note that for experiments in 2009 14C-assimilation by the JTB255 clade was not measured (for details, see Supplementary Information).
Although they can be important autotrophs in organic-poor deep-sea sediments (Molari et al., 2013) nitrifying Archaea have a minor role in dark carbon fixation in the sediments we measured. In our study, Archaea occurred at low relative abundances and assimilated less 14C than Gammaproteobacteria (Figures 4a and b and Supplementary Figure S7).
Genomics suggests thioautotrophy in uncultured Gammaproteobacteria
We previously showed that key genes of sulfur oxidation in Janssand sediments are mainly affiliated with Gammaproteobacteria (Lenk et al., 2011). To further investigate the metabolism of the candidate chemolithoautotrophic Gammaproteobacteria, we sequenced the amplified genomic DNA of a single cell of the SSr clade from Janssand sediment. In addition, we recovered a metagenomic bin of a member of the Acidiferrobacter clade from a deep-sea hydrothermal chimney that displayed 91% sequence identity to 16S rRNA gene sequences from site Janssand (Supplementary Figure S4).
The assembled SAG of the SSr cell (‘WSgam209’) consists of 1.9 Mbp on 311 scaffolds (Supplementary Table S3). In addition to the cytochrome c oxidase for oxygen respiration, the genome encoded a reverse dissimilatory adenosine-5′-phosphosulfate (APS)-reductase subunit A (AprA), a widely distributed enzyme catalyzing the oxidation of sulfite to sulfate (Meyer and Kuever, 2007). Similar to the 16S rRNA gene also the AprA is affiliated with the sulfur-oxidizing symbionts of Oligobrachia haakonmosbiensis (Supplementary Figures S4 and S8), indicating a congruent phylogeny of both phylogenetic markers. Moreover, it encoded the large and small subunits of the RuBisCO form I (CbbL, CbbS) (Supplementary Table S5).
The metagenomic bin of the Acidiferrobacter-clade organism (‘Acidiferrobacter-a7’) consists of 2 Mbp on 66 scaffolds (Supplementary Table S3). It contained genes for thiosulfate oxidation (Sox-multienzyme complex, soxABXYZ), sulfite oxidation (soeABC) and carbon fixation (RuBisCO form I subunits cbbL and cbbS) (Supplementary Table S4). Together with the measured carbon fixation of SSr and the Acidiferrobacter clades at site Janssand, the identification of sulfur oxidation and carbon fixation genes in both clades provides the genetic background of their chemolithoautotrophic potential.
Metatranscriptomics underscores the role of Gammaproteobacteria in thioautotrophy
To further test, whether Gammaproteobacteria in these sediments are the major active chemolithoautotrophs, we sequenced triplicate metatranscriptomes from the sulfide transition zone of Janssand sediments. Transcript reads were mapped to reference sequences from the GenBank database, functional gene libraries, metagenomic fragments and the SSr-single cell genome recovered from site Janssand (this study; Lenk et al., 2011, 2012).
To identify the expressed carbon fixation pathways, we mapped the metatranscriptomic reads to gene sequences of RuBisCO form I and form II (cbbL, cbbM) and of ATP citrate lyase (aclAB) from cultured representatives and environmental sequences from diverse aquatic environments including sediments. All transcripts of RuBisCO genes, on average 112 reads, mapped to those of sulfur-oxidizing Gammaproteobacteria, confirming active carbon fixation through the CBB-cycle in Janssand sediments. Only very few transcripts (n⩽6) could be mapped on genes encoding an epsilonproteobacterial ATP citrate lyase (Supplementary Table S4), reflecting the low relative abundance and low activity of sulfur-oxidizing Epsilonproteobacteria in these sediments (Figure 2; Lenk et al., 2011).
To identify the expressed sulfur oxidation pathways and the respective organisms in the sulfur transition zone in Janssand sediments, we mapped the metatranscriptomic reads to references databases containing gene sequences encoding SoxB, AprA and subunits of the reverse dissimilatory sulfite reductase, DsrAB. The major fraction (69–82%) of transcripts mapped to dsrAB, soxB and aprA sequences that are affiliated with Gammaproteobacteria (Figure 6 and Supplementary Figure S8), further supporting their central role in chemolithoautotrophy. Here, dsrAB transcripts were fourfold higher than soxB transcripts. The identification of the sulfur oxidation pathway prevailing in marine sediments has important implications for modeling of carbon budgets, as the reverse Dsr (rDsr) pathway may allow a higher ATP gain that can be used for carbon fixation than the complete Sox-multienzyme complex (Klatt and Polerecky, 2015). In the rDsr pathway electrons finally enter the electron transport chain at the level of quinone (Holkenbrink et al., 2011), while the Sox-multienzyme complex donates electrons to cytochrome c (Kelly et al., 1997), probably resulting in higher energy yields for rDsr-encoding sulfur oxidizers. However, further in situ studies are essential to confirm this observation.
Taxonomic affiliation of metatranscriptomic reads of key genes from sulfur and hydrogen oxidation pathways. Metatranscriptomic reads from the sulfide transition zone (Janssand surface sediments 2–3 cmbsf) were mapped to key genes of sulfur and hydrogen oxidation genes (aprA, adenosine-5′-phosphosulfate reductase; dsrAB, reverse dissimilatory sulfite reductase; soxB, Sox-multienzyme system; uptake [NiFe]-hydrogenase). Metatranscriptomes were sequenced in triplicates, and the average values are displayed. Transcript abundance is normalized for gene length and number of reads per data set.
Very few reads mapped to bacterial (n⩽2) and archaeal (n⩽5) amoA genes encoding the ammonia monooxygenase subunit A (AmoA) (Supplementary Table S1). This is consistent with the minor role of Archaea in carbon assimilation measured in our study (Supplementary Figure S7), and is also consistent with the very low nitrification rates measured at site Janssand (Marchant et al., 2014). Overall our data confirm previous biogeochemical models suggesting a low impact of nitrification on chemolithoautotrophic production in coastal sediments (Middelburg, 2011; Boschker et al., 2014).
Hydrogen is likely an alternative energy source for sulfur-oxidizing Gammaproteobacteria
Recently, uptake [NiFe]-hydrogenase genes were found in metagenomic bins of Gammaproteobacteria from estuarine sediments, indicating alternative energy sources for dark carbon fixation under oxic to suboxic conditions (Baker et al., 2015). As the ability to oxidize hydrogen has been shown to confer metabolic plasticity to symbiotic and pelagic sulfur-oxidizing bacteria (Petersen et al., 2011; Anantharaman et al., 2013; Hansen and Perner, 2015), we tested whether hydrogen could serve as an alternative energy source also for gammaproteobacterial sulfur oxidizers in marine sediments, for example, to respire sulfur under anoxic conditions (Laurinavichene et al., 2007). To overcome the lack of reference sequences from marine sediments, we constructed an uptake [NiFe]-hydrogenase gene library from Janssand sediments. The recovered hydrogenase gene diversity comprised different physiological groups from phyla such as Proteobacteria and Bacteroidetes (Supplementary Figure S9). A substantial fraction (40%) of all hydrogenase transcripts were assigned to sequences of diverse Gammaproteobacteria, in particular to those grouping with sulfur-oxidizing bacteria (Figure 6 and Supplementary Figure S9). Expression levels of hydrogenase genes were lower than those of sulfur oxidation genes, but were in the same order of magnitude (Figure 6).
To link a potential hydrogen-oxidizing activity with sulfur-oxidizing Gammaproteobacteria, we searched for co-localization and co-expression of key genes of both pathways. First, we identified a metagenomic fragment from Janssand, affiliated with Gammaproteobacteria, which encoded both the uptake [NiFe]-hydrogenase HupSL and the rDsr operon (Supplementary Figure S10). Notably, the SAG of the SSr-group recovered from site Janssand also encodes an uptake [NiFe]-hydrogenase gene in addition to aprA and cbbLS. Moreover, these genes were among the top 20 transcribed genes out of 2008 identified genes (Supplementary Figure S11).
Collectively, our single-cell genomic, metagenomic and metatranscriptomic data indicate that Gammaproteobacteria in marine surface sediments may use both reduced sulfur species and hydrogen as energy sources for carbon fixation. In fact, the thioautotroph Sulfurimonas denitrificans grows more efficiently with hydrogen than with thiosulfate, when the electron acceptor nitrate is limiting (Han and Perner, 2014). Hence, hydrogen oxidation could be a hitherto overlooked energy source for carbon fixation in marine sediments.
Key functions of ubiquitous chemolithoautotrophic Gammaproteobacteria in sediments
Overall, our molecular and 14C-assimilation data suggest that rather than a single group, a stable assemblage of Gammaproteobacteria drives dark carbon fixation in coastal surface sediments. In particular, we showed that members of the Acidiferrobacter, the JTB255 and the SSr clade occur in sediments worldwide and fix carbon in rates similar to those of uncultured sulfur-oxidizing bacteria from other aquatic habitats. Our genomic and metatranscriptomic evidence supports the previous assumption that chemolithoautotrophy in marine surface sediments is mainly driven by sulfur oxidation (Middelburg, 2011). However, the expression of uptake [NiFe]-hydrogenases in the SSr clade and other Gammaproteobacteria suggests that these organisms may also use hydrogen as an energy source for carbon fixation. On the basis of our data, we cannot exclude the possibility that other chemolithoautotrophic pathways such as ferrous iron oxidation also contributed to dark carbon fixation, but these are probably minor in organic- and sulfide-rich systems. Molecular, isotopic and physiological studies will be critical to determine, how other chemoautotrophic processes such as nitrification, metal oxidation, sulfur disproportionation (Jørgensen, 1990) and possibly also hydrogen-dependent sulfate respiration (Boschker et al., 2014) contribute to dark carbon fixation in marine sediments. Intriguingly, sulfur-oxidizing, autotrophic Gammaproteobacteria including members of the SSr clade were recently shown to be associated with heterotrophic, electrogenic ‘cable bacteria’. These were hypothesized to use cable bacteria as an electron sink during autotrophic sulfur oxidation (Vasquez-Cardenas et al., 2015). Considering their cosmopolitan distribution, their metabolic lifestyle and their ecological importance, the Acidiferrobacter-, the JTB255-, and the three symbiont-related clades may be benthic counterparts to the gammaproteobacterial SUP05 clade, key organisms for sulfur and carbon cycling in hydrothermal plumes and OMZs (Canfield et al., 2010; Wright et al., 2012; Anantharaman et al., 2013; Glaubitz et al., 2013).
Role of chemolithoautotrophic gammaproteobacteria in carbon cycling
Coastal sediments are global hot spots of carbon cycling. The importance of marine vegetation such as sea grass, salt marshes and mangroves for carbon sequestration is already well established (Duarte et al., 2005), but the role of coastal sediments as hot spots of microbial dark carbon fixation was only recently realized (Middelburg, 2011). According to our most conservative estimate, 70% of dark carbon fixation in coastal sediments are driven by chemolithoautotrophic Gammaproteobacteria (Figure 5). These could fix 122 Tg C/year in Earth's coastal sediments (assuming a total of 175 Tg/year, from Middelburg 2011), which is similar to the 111 Tg carbon buried yearly by marine vegetated habitats worldwide (Duarte et al., 2005). It is still unclear whether significant amounts of carbon fixed in the dark are buried into subsurface sediments. However, because identical/almost identical chemolithoautotrophic Gammaproteobacteria are frequently found in surface and subsurface sediments, they may have the potential to trap inorganic carbon and survive for centuries in subsurface sediments by tapping yet unknown sources of energy. Understanding whether the buried populations are in a state of dynamic equilibrium or whether they merely survive will be essential for assessing their role as a carbon sink.
Even though chemolithoautotrophy in shelf sediments mostly represents a ‘secondary production’, as it is ultimately based on energy from recycled organic matter (Middelburg, 2011), it may mitigate carbon (and sulfide) emissions from re-mineralized organic matter already at sediment surfaces. As marine sediments are the main site of global carbon sequestration, it is imperative to understand the processes and microorganisms that govern rates of burial of organic and inorganic carbon in these habitats. Here, our study provides first detailed insights into the microbiology of a largely overlooked aspect of the marine carbon cycle and highlights the environmental importance of widely distributed chemolithoautotrophic, most likely sulfur-oxidizing Gammaproteobacteria. As hypoxic events will expand and intensify in a warming ocean, sulfur-dependent carbon cycling will be more prevalent not only in pelagic OMZ, but also in organic-rich coastal sediments. Thus, sulfur-oxidizing and carbon-fixing microorganisms may have an increasingly important role in attenuating the rising emissions of sulfide and inorganic carbon to ocean waters and ultimately to the atmosphere.
Accession codes
References
Alonso C, Pernthaler J . (2005). Incorporation of glucose under anoxic conditions by bacterioplankton from coastal North Sea surface waters. Appl Environ Microbiol 71: 1709–1716.
Anantharaman K, Breier JA, Sheik CS, Dick GJ . (2013). Evidence for hydrogen oxidation and metabolic plasticity in widespread deep-sea sulfur-oxidizing bacteria. Proc Natl Acad Sci USA 110: 330–335.
Baker BJ, Lazar CS, Teske AP, Dick GJ . (2015). Genomic resolution of linkages in carbon, nitrogen, and sulfur cycling among widespread estuary sediment bacteria. Microbiome 3: 14.
de Beer D, Wenzhöfer F, Ferdelman TG, Boehme SE, Huettel M, van Beusekom JEE et al. (2005). Transport and mineralization rates in North Sea sandy intertidal sediments, Sylt-Rømø Basin, Wadden Sea. Limnol Oceanogr 50: 113–127.
Billerbeck M, Werner U, Polerecky L, Walpersdorf E, deBeer D, Huettel M . (2006). Surficial and deep pore water circulation governs spatial and temporal scales of nutrient recycling in intertidal sand flat sediment. Mar Ecol Prog Ser 326: 61–76.
Boschker HTS, Nold SC, Wellsbury P, Bos D, de Graaf W, Pel R et al. (1998). Direct linking of microbial populations to specific biogeochemical processes by 13C-labelling of biomarkers. Nature 392: 801–805.
Boschker HTS, Vasquez-Cardenas D, Bolhuis H, Moerdijk-Poortvliet TWC, Moodley L . (2014). Chemoautotrophic carbon fixation rates and active bacterial communities in intertidal marine sediments. PLoS ONE 9: e101443.
Bowman JP, McCammon SA, Dann AL . (2005). Biogeographic and quantitative analyses of abundant uncultivated γ-proteobacterial clades from marine sediment. Microb Ecol 49: 451–460.
Bowman JP, McCuaig RD . (2003). Biodiversity, community structural shifts, and biogeography of prokaryotes within Antarctic continental shelf sediment. Appl Environ Microbiol 69: 2463–2483.
Burdige DJ . (2007). Preservation of organic matter in marine sediments: controls, mechanisms, and an imbalance in sediment organic carbon budgets? Chem Rev 107: 467–485.
Canfield DE, Stewart FJ, Thamdrup B, Brabandere LD, Dalsgaard T, Delong EF et al. (2010). A cryptic sulfur cycle in oxygen-minimum-zone waters off the Chilean coast. Science 330: 1375–1378.
Cleland D, Krader P, McCree C, Tang J, Emerson D . (2004). Glycine betaine as a cryoprotectant for prokaryotes. J Microbiol Methods 58: 31–38.
Cline JD . (1969). Spectrophotometric determination of hydrogen sulfide in natural waters. Limnol Oceanogr 14: 454–458.
Duarte CM, Middelburg JJ, Caraco N . (2005). Major role of marine vegetation on the oceanic carbon cycle. Biogeosciences 2: 1–8.
Emil Ruff S, Probandt D, Zinkann A-C, Iversen MH, Klaas C, Würzberg L et al. (2014). Indications for algae-degrading benthic microbial communities in deep-sea sediments along the Antarctic Polar Front. Deep Sea Res Part II Top Stud Oceanogr 108: 6–16.
Gittel A, Mußmann M, Sass H, Cypionka H, Könneke M . (2008). Identity and abundance of active sulfate-reducing bacteria in deep tidal flat sediments determined by directed cultivation and CARD-FISH analysis. Environ Microbiol 10: 2645–2658.
Glaubitz S, Kießlich K, Meeske C, Labrenz M, Jürgens K . (2013). SUP05 dominates the gammaproteobacterial sulfur oxidizer assemblages in pelagic redoxclines of the central Baltic and Black Seas. Appl Environ Microbiol 79: 2767–2776.
Gobet A, Böer SI, Huse SM, van Beusekom JEE, Quince C, Sogin ML et al. (2012). Diversity and dynamics of rare and of resident bacterial populations in coastal sands. ISME J 6: 542–553.
Grote J, Schott T, Bruckner CG, Glöckner FO, Jost G, Teeling H et al. (2012). Genome and physiology of a model Epsilonproteobacterium responsible for sulfide detoxification in marine oxygen depletion zones. Proc Natl Acad Sci USA 109: 506–510.
Hallberg KB, Hedrich S, Johnson DB . (2011). Acidiferrobacter thiooxydans, gen. nov. sp. nov.; an acidophilic, thermo-tolerant, facultatively anaerobic iron- and sulfur-oxidizer of the family Ectothiorhodospiraceae. Extremophiles 15: 271–279.
Han Y, Perner M . (2014). The role of hydrogen for Sulfurimonas denitrificans’ metabolism. PLoS One 9: e106218.
Hansen M, Perner M . (2015). A novel hydrogen oxidizer amidst the sulfur-oxidizing Thiomicrospira lineage. ISME J 9: 696–707.
Hawley AK, Brewer HM, Norbeck AD, Paša-Tolić L, Hallam SJ . (2014). Metaproteomics reveals differential modes of metabolic coupling among ubiquitous oxygen minimum zone microbes. Proc Natl Acad Sci USA 111: 11395–11400.
Hedges JI, Keil RG . (1995). Sedimentary organic matter preservation: an assessment and speculative synthesis. Mar Chem 49: 81–115.
Herlemann DP, Labrenz M, Jürgens K, Bertilsson S, Waniek JJ, Andersson AF . (2011). Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J 5: 1571–1579.
Holkenbrink C, Barbas SO, Mellerup A, Otaki H, Frigaard N-U . (2011). Sulfur globule oxidation in green sulfur bacteria is dependent on the dissimilatory sulfite reductase system. Microbiology 157: 1229–1239.
Hügler M, Sievert SM . (2011). Beyond the Calvin cycle: autotrophic carbon fixation in the ocean. Annu Rev Mar Sci 3: 261–289.
Hunter EM, Mills HJ, Kostka JE . (2006). Microbial community diversity associated with carbon and nitrogen cycling in permeable shelf sediments. Appl Environ Microbiol 72: 5689–5701.
Jansen S, Walpersdorf E, Werner U, Billerbeck M, Böttcher ME, de Beer D . (2009). Functioning of intertidal flats inferred from temporal and spatial dynamics of O2, H2S and pH in their surface sediment. Ocean Dyn 59: 317–332.
Jørgensen BB . (1990). A thiosulfate shunt in the sulfur cycle of marine sediments. Science 249: 152–154.
Jørgensen BB . (2011). Deep subseafloor microbial cells on physiological standby. Proc Natl Acad Sci USA 108: 18193–18194.
Jost G, Zubkov MV, Yakushev E, Labrenz M, Jürgens K . (2008). High abundance and dark CO2 fixation of chemolithoautotrophic prokaryotes in anoxic waters of the Baltic Sea. Limnol Oceanogr 53: 14–22.
Kelly DP, Shergill JK, Lu W-P, Wood AP . (1997). Oxidative metabolism of inorganic sulfur compounds by bacteria. Antonie Van Leeuwenhoek 71: 95–107.
Kim B-S, Kim BK, Lee J-H, Kim M, Lim YW, Chun J . (2008). Rapid phylogenetic dissection of prokaryotic community structure in tidal flat using pyrosequencing. J Microbiol 46: 357–363.
Klatt JM, Polerecky L . (2015). Assessment of the stoichiometry and efficiency of CO2 fixation coupled to reduced sulfur oxidation. Aquat Microbiol 6: 484.
Kleiner M, Wentrup C, Lott C, Teeling H, Wetzel S, Young J et al. (2012). Metaproteomics of a gutless marine worm and its symbiotic microbial community reveal unusual pathways for carbon and energy use. Proc Natl Acad Sci USA 109: E1173–E1182.
Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M et al. (2012). Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res 41: e1.
Langmead B, Salzberg SL . (2012). Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359.
Laurinavichene TV, Rákhely G, Kovács KL, Tsygankov AA . (2007). The effect of sulfur compounds on H2 evolution/consumption reactions, mediated by various hydrogenases, in the purple sulfur bacterium Thiocapsa roseopersicina. Arch Microbiol 188: 403–410.
Lavik G, Stührmann T, Brüchert V, Van der Plas A, Mohrholz V, Lam P et al. (2009). Detoxification of sulphidic African shelf waters by blooming chemolithotrophs. Nature 457: 581–584.
Lee N, Nielsen PH, Andreasen KH, Juretschko S, Nielsen JL, Schleifer KH et al. (1999). Combination of fluorescent in situ hybridization and microautoradiography - a new tool for structure-function analyses in microbial ecology. Appl Environ Microbiol 65: 1289–1297.
Lenk S, Arnds J, Zerjatke K, Musat N, Amann R, Mußmann M . (2011). Novel groups of Gammaproteobacteria catalyse sulfur oxidation and carbon fixation in a coastal, intertidal sediment. Environ Microbiol 13: 758–774.
Lenk S, Moraru C, Hahnke S, Arnds J, Richter M, Kube M et al. (2012). Roseobacter clade bacteria are abundant in coastal sediments and encode a novel combination of sulfur oxidation genes. ISME J 6: 2178–2187.
Li W . (1982). Estimating heterotrophic bacterial productivity by inorganic radiocarbon uptake - importance of establishing time courses of uptake. Mar Ecol Prog Ser 8: 167–172.
Liu J, Liu X, Wang M, Qiao Y, Zheng Y, Zhang X-H . (2014). Bacterial and archaeal communities in sediments of the north Chinese marginal seas. Microb Ecol 70: 105–117.
Lösekann T, Robador A, Niemann H, Knittel K, Boetius A, Dubilier N . (2008). Endosymbioses between bacteria and deep-sea siboglinid tubeworms from an Arctic Cold Seep (Haakon Mosby Mud Volcano, Barents Sea). Environ Microbiol 10: 3237–3254.
Manefield M, Whiteley AS, Griffiths RI, Bailey MJ . (2002). RNA stable isotope probing, a novel means of linking microbial community function to phylogeny. Appl Environ Microbiol 68: 5367–5373.
Marchant HK, Lavik G, Holtappels M, Kuypers MMM . (2014). The fate of nitrate in intertidal permeable sediments. PLoS ONE 9: e104517.
Markowitz VM, Chen I-MA, Palaniappan K, Chu K, Szeto E, Grechkin Y et al. (2012). IMG: the integrated microbial genomes database and comparative analysis system. Nucleic Acids Res 40: D115–D122.
Mattes TE, Nunn BL, Marshall KT, Proskurowski G, Kelley DS, Kawka OE et al. (2013). Sulfur oxidizers dominate carbon fixation at a biogeochemical hot spot in the dark ocean. ISME J 7: 2349–2360.
Meyer B, Kuever J . (2007). Molecular analysis of the diversity of sulfate-reducing and sulfur-oxidizing prokaryotes in the environment, using aprA as functional marker gene. Appl Environ Microbiol 73: 7664–7679.
Middelburg JJ . (2011). Chemoautotrophy in the ocean. Geophys Res Lett 38: 94–97.
Molari M, Manini E, Dell’Anno A . (2013). Dark inorganic carbon fixation sustains the functioning of benthic deep-sea ecosystems. Glob Biogeochem Cycles 27: 212–221.
Musat N, Halm H, Winterholler B, Hoppe P, Peduzzi S, Hillion F et al. (2008). A single-cell view on the ecophysiology of anaerobic phototrophic bacteria. Proc Natl Acad Sci USA 105: 17861–17866.
Muyzer G, Brinkhoff T, Nuebel U, Santegoeds C, Schaefer H, Waver C . (1998). Denaturing gradient gel electrophoresis (DGGE) in microbial ecology. In: Akkermans ADL, van Elsas JD, de Bruijn FJ (ed). Molecular Microbial Ecology Manual. Kluwer Academic Publishers: Dordrecht, The Netherlands, pp 1–27.
Orcutt BN, Sylvan JB, Knab NJ, Edwards KJ . (2011). Microbial ecology of the dark ocean above, at, and below the seafloor. Microbiol Mol Biol Rev 75: 361–422.
Parkes RJ, Cragg BA, Getliff JM, Harvey SM, Fry JC, Lewis CA et al. (1993). A quantitative study of microbial decomposition of biopolymers in Recent sediments from the Peru Margin. Mar Geol 113: 55–66.
Pernthaler A, Pernthaler J, Amann R . (2002). Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl Environ Microbiol 68: 3094–3101.
Petersen JM, Zielinski FU, Pape T, Seifert R, Moraru C, Amann R et al. (2011). Hydrogen is an energy source for hydrothermal vent symbioses. Nature 476: 176–180.
Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J et al. (2007). SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35: 7188–7196.
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41: D590–D596.
Radajewski S, Ineson P, Parekh NR, Murrell JC . (2000). Stable-isotope probing as a tool in microbial ecology. Nature 403: 646–649.
Reinthaler T, van Aken HM, Herndl GJ . (2010). Major contribution of autotrophy to microbial carbon cycling in the deep North Atlantic’s interior. Deep Sea Res Part II Top Stud Oceanogr 57: 1572–1580.
Rinke C, Schmitz-Esser S, Stoecker K, Nussbaumer AD, Molnár DA, Vanura K et al. (2006). ‘Candidatus Thiobios zoothamnicoli,’ an ectosymbiotic bacterium covering the giant marine ciliate Zoothamnium niveum. Appl Environ Microbiol 72: 2014–2021.
Roslev P, Larsen MB, Jorgensen D, Hesselsoe M . (2004). Use of heterotrophic CO2 assimilation as a measure of metabolic activity in planktonic and sessile bacteria. J Microbiol Methods 59: 381–393.
Røy H, Lee JS, Jansen S, de Beer D . (2008). Tide-driven deep pore-water flow in intertidal sand flats. Limnol Oceanogr 53: 1521–1530.
Ruff SE, Biddle JF, Teske AP, Knittel K, Boetius A, Ramette A . (2015). Global dispersion and local diversification of the methane seep microbiome. Proc Natl Acad Sci USA 112: 4015–4020.
Salman V, Bailey JV, Teske A . (2013). Phylogenetic and morphologic complexity of giant sulphur bacteria. Antonie Van Leeuwenhoek 104: 169–186.
Schauer R, Bienhold C, Ramette A, Harder J . (2009). Bacterial diversity and biogeography in deep-sea surface sediments of the South Atlantic Ocean. ISME J 4: 159–170.
Schippers A, Neretin LN, Kallmeyer J, Ferdelman TG, Cragg BA, John Parkes R et al. (2005). Prokaryotic cells of the deep sub-seafloor biosphere identified as living bacteria. Nature 433: 861–864.
Seidel M, Graue J, Engelen B, Köster J, Sass H, Rullkötter J . (2012). Advection and diffusion determine vertical distribution of microbial communities in intertidal sediments as revealed by combined biogeochemical and molecular biological analysis. Org Geochem 52: 114–129.
Sekar R, Fuchs BM, Amann R, Pernthaler J . (2004). Flow sorting of marine bacterioplankton after fluorescence in situ hybridization. Appl Environ Microbiol 70: 6210–6219.
Siyambalapitiya N, Blackall LL . (2005). Discrepancies in the widely applied GAM42a fluorescence in situ hybridisation probe for Gammaproteobacteria. FEMS Microbiol Lett 242: 367–373.
Swan BK, Martinez-Garcia M, Preston CM, Sczyrba A, Woyke T, Lamy D et al. (2011). Potential for chemolithoautotrophy among ubiquitous bacteria lineages in the dark ocean. Science 333: 1296–1300.
Thomas F, Giblin AE, Cardon ZG, Sievert SM . (2014). Rhizosphere heterogeneity shapes abundance and activity of sulfur-oxidizing bacteria in vegetated salt marsh sediments. Front Microbiol 5: 1–14.
Vasquez-Cardenas D, van de Vossenberg J, Polerecky L, Malkin SY, Schauer R, Hidalgo-Martinez S et al. (2015). Microbial carbon metabolism associated with electrogenic sulphur oxidation in coastal sediments. ISME J 9: 1966–1978.
Wang L, Liu L, Zheng B, Zhu Y, Wang X . (2013). Analysis of the bacterial community in the two typical intertidal sediments of Bohai Bay, China by pyrosequencing. Mar Pollut Bull 72: 181–187.
Wood HG, Werkman CH . (1936). The utilisation of CO2 in the dissimilation of glycerol by the propionic acid bacteria. Biochem J 30: 48–53.
Woyke T, Teeling H, Ivanova NN, Huntemann M, Richter M, Gloeckner FO et al. (2006). Symbiosis insights through metagenomic analysis of a microbial consortium. Nature 443: 950–955.
Wright JJ, Konwar KM, Hallam SJ . (2012). Microbial ecology of expanding oxygen minimum zones. Nat Rev Microbiol 10: 381–394.
Zerjatke K . (2009). Mikroautoradiographie und Genexpression von Bakterien im Oberflächensediment der Gezeitenzone des Wattenmeeres. Diploma thesis. University of Bremen: Bremen, Germany.
Zheng B, Wang L, Liu L . (2014). Bacterial community structure and its regulating factors in the intertidal sediment along the Liaodong Bay of Bohai Sea, China. Microbiol Res 169: 585–592.
Zhou J, Bruns MA, Tiedje JM . (1996). DNA recovery from soils of diverse composition. Appl Environ Microbiol 62: 316–322.
Ziehe D . (2009) Aminosäure-D/L-Verhältnisse in biogenen Carbonaten als Schlüssel zur Datierung holozäner Sedimentationsvorgänge im norddeutschen Küstenraum PhD thesis. University of Oldenburg: Oldenburg, Germany.
Zubkov MV, Fuchs BM, Tarran GA, Burkill PH, Amann R . (2003). High rate of uptake of organic nitrogen compounds by Prochlorococcus cyanobacteria as a key to their dominance in oligotrophic oceanic waters. Appl Environ Microbiol 69: 1299–1304.
Acknowledgements
We greatly acknowledge the crew of R/V Navicula from the ICBM Oldenburg for ship time and assistance. We thank the crew of the R/V Meteor and R/V Sonne and the ROV team of the MARUM Quest 4000 m. We greatly acknowledge the chief scientist Wolfgang Bach for his excellent support during cruise SO216 with R/V Sonne, which was an integral part of the Cluster of Excellence of the MARUM ‘The Ocean in the Earth System, Research Area GB: Geosphere-Biosphere Interactions’ funded by the German Research Foundation (DFG). The cruise SO216 was funded by a grant (03G0216) from the Bundesministerium für Bildung und Forschung (BMBF) awarded to Wolfgang Bach and co-PIs. RS was supported by the US National Science Foundation grant OCE-1232982. We thank Falk Warnecke for support in sediment sampling and for sequencing of the subsurface samples. Special thanks go to Rudolf Amann for helpful discussions and continuous support. This study contributes to the project ABYSS funded by the European Research Council Advanced Investigator Grant 294757 to Antje Boetius. Further funding was provided by the Max Planck Society.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Rights and permissions
About this article
Cite this article
Dyksma, S., Bischof, K., Fuchs, B. et al. Ubiquitous Gammaproteobacteria dominate dark carbon fixation in coastal sediments. ISME J 10, 1939–1953 (2016). https://doi.org/10.1038/ismej.2015.257
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2015.257
This article is cited by
-
Diversity and potential host-interactions of viruses inhabiting deep-sea seamount sediments
Nature Communications (2024)
-
Simultaneous sulfate and nitrate reduction in coastal sediments
ISME Communications (2023)
-
Microbial community and predictive functionalities associated with the marine sediment of Coastal Gujarat
Environmental Science and Pollution Research (2023)
-
Microbial diversity and functional profiling in coastal tidal flat sediment with pollution of nutrients and potentially toxic elements
Journal of Soils and Sediments (2023)
-
Microbial community dynamics and its correlation with environmental factors in the water of polyculture ponds containing Penaeus japonicus, Portunus trituberculatus and Sinonovacula constricta
Aquatic Ecology (2023)