Abstract
Corals experience intimate associations with distinct populations of marine microorganisms, but the microbial behaviours underpinning these relationships are poorly understood. There is evidence that chemotaxis is pivotal to the infection process of corals by pathogenic bacteria, but this evidence is limited to experiments using cultured isolates under laboratory conditions. We measured the chemotactic capabilities of natural populations of coral-associated bacteria towards chemicals released by corals and their symbionts, including amino acids, carbohydrates, ammonium and dimethylsulfoniopropionate (DMSP). Laboratory experiments, using a modified capillary assay, and in situ measurements, using a novel microfabricated in situ chemotaxis assay, were employed to quantify the chemotactic responses of natural microbial assemblages on the Great Barrier Reef. Both approaches showed that bacteria associated with the surface of the coral species Pocillopora damicornis and Acropora aspera exhibited significant levels of chemotaxis, particularly towards DMSP and amino acids, and that these levels of chemotaxis were significantly higher than that of bacteria inhabiting nearby, non-coral-associated waters. This pattern was supported by a significantly higher abundance of chemotaxis and motility genes in metagenomes within coral-associated water types. The phylogenetic composition of the coral-associated chemotactic microorganisms, determined using 16S rRNA amplicon pyrosequencing, differed from the community in the seawater surrounding the coral and comprised known coral associates, including potentially pathogenic Vibrio species. These findings indicate that motility and chemotaxis are prevalent phenotypes among coral-associated bacteria, and we propose that chemotaxis has an important role in the establishment and maintenance of specific coral–microbe associations, which may ultimately influence the health and stability of the coral holobiont.
Similar content being viewed by others
Introduction
Corals host bacterial communities that are phylogenetically distinct, more active and more abundant than the bacterial communities in the surrounding seawater (Ducklow and Mitchell, 1979a; Paul et al., 1986; Rohwer et al., 2001, 2002; Frias-Lopez et al., 2002; Kellogg, 2004; Rosenberg et al., 2007; Sweet et al., 2011). Although the recent application of molecular techniques has begun to unravel the complex nature of coral–bacteria interactions (Rohwer et al., 2002; Rosenberg et al., 2007; Ceh et al., 2011, 2012), we only have a rudimentary understanding of the ecological mechanisms and bacterial behaviours underpinning these ecological associations. Recent work focussed on coral pathogens has revealed that chemotaxis may be a potentially important phenotype among coral-associated bacteria (Banin et al., 2001; Rosenberg et al., 2007; Meron et al., 2009; Vega Thurber et al., 2009; Garren et al., 2014).
Chemotaxis may be a particularly beneficial phenotype within reefs because the coral surface microenvironment is characterised by strong gradients of chemical cues and organic material (Kuhl et al., 1995; Mass et al., 2010). Coral mucus and the exudates of the symbiotic dinoflagellate Symbiodinium sp. are highly enriched in microbial growth substrates including amino acids, carbohydrates and the organic sulphur compound dimethylsulfoniopropionate (DMSP) (Von Holt and Von Holt, 1968; Ducklow and Mitchell, 1979b; Meikle et al., 1988; Wild et al., 2005, 2010; Raina et al., 2009, 2010, 2013; Garren et al., 2014), and gradients of these materials extend from the coral surface into the immediately surrounding seawater (Garren et al., 2014). The capacity to employ chemotaxis to exploit the resource-rich or infochemical-rich coral surface microenvironment may thus provide considerable advantages for reef microorganisms by enabling access to limiting substrates or potential animal hosts.
Marine bacteria exhibit high-performance motility (Mitchell et al., 1995, 1996; Grossart et al., 2001) and chemotaxis (Stocker et al., 2008; Seymour et al., 2009; Stocker and Seymour, 2012), and there is evidence that bacterial chemotaxis may be an ecologically important phenotype within coral reefs (Banin et al., 2001; Rosenberg et al., 2007; Meron et al., 2009; Vega Thurber et al., 2009; Garren et al., 2014). Some of the earliest work on marine bacterial chemotaxis demonstrated that coral and Symbiodinium exudates are potent chemoattractants (Chet and Mitchell, 1976; Bartlett and Matsumura, 1986), and chemotaxis and motility are important phenotypes for the coral pathogens Vibrio shiloi and V. coralliilyticus to locate, invade and colonise their coral hosts (Banin et al., 2001; Koren and Rosenberg, 2006; Rosenberg et al., 2007; Meron et al., 2009; Kimes et al., 2011). Recently, it has been demonstrated that V. coralliilyticus exhibits extremely strong chemotactic responses towards DMSP to locate heat-stressed colonies of its coral host, Pocillopora damicornis (Garren et al., 2014).
Although increasing evidence suggests that chemotaxis is an important phenotype among marine bacterial populations (Blackburn et al., 1998; Stocker et al., 2008; Stocker and Seymour, 2012), most research in this area has relied on the use of cultured isolates and experiments performed under laboratory conditions. Yet, the majority of marine bacteria are not amenable to cultivation, excluding potentially important representatives of natural bacterial communities from laboratory experiments (Jannasch and Jones, 1959; Hoppe, 1976; Bianchi and Giuliano, 1996). In addition, it is important to establish the extent to which swimming and chemosensory capabilities of laboratory isolates reflect their natural state, and how isolates respond to chemoattractants in the presence of natural populations. Here, we aim to expand our understanding of coral–microbe interactions by examining chemotaxis among natural populations of coral-associated bacteria using a combination of laboratory-based and in situ experiments.
Materials and methods
This study was conducted on Heron Island in the Capricorn Bunker Group on the southern Great Barrier Reef, Australia (23°26′S, 151°54′E) during two consecutive winter sampling seasons in July 2010 and July 2011.
Laboratory chemotaxis experiments
To quantify the level of chemotaxis demonstrated by natural communities of coral reef bacteria, we performed a set of laboratory-based studies using seawater samples collected from coral-associated and nearby non-coral-associated environments. Seawater (1 l) was collected from two environments: (i) by placing the mouth of a sterile 1-l Schott bottle immediately adjacent (<1 cm distance) to the surface of colonies of the coral species Pocillopora damicornis, in 1.5 m depth within the reef of Heron Island (23°26′28′′S, 151°55′11′′E) (‘coral associated’) (Supplementary Figure S1) and (ii) at the surface of deeper (10 m depth) open water, outside of the Heron Island reef overlaying a large patch of sandy substrate, where no corals were present within a radius of 10 m (23°26′04′′S, 151°55′20′′E) (‘non-coral associated’) (Supplementary Figure S1). Water samples were returned to the Heron Island Research Station laboratory and used immediately (within 10 min) for chemotaxis experiments.
In these laboratory experiments, we examined chemotactic responses using a modified version of the capillary assay (Pfeffer, 1884; Adler, 1973), whereby sterile 1-ml syringes were filled with 150 μl of putative chemoattractant. Syringes were inserted into 100-ml vials containing seawater collected from the environment (Dennis et al., 2013). Each of three replicates involved a single syringe being inserted into one vial of seawater. Once placed into the vial of seawater chemotattractants diffused out of the syringe into the seawater suspension, and chemotactic bacteria within the seawater migrated into the syringe. Putative chemoattractants were selected according to their relevance as components of coral mucus (Von Holt and Von Holt, 1968; Muscatine and Cernichiari, 1969; Ducklow and Mitchell, 1979b; Meikle et al., 1988; Hill et al., 1995; Fitzgerald and Szmant, 1997; Broadbent et al., 2002; Broadbent and Jones, 2004; Wild et al., 2005, 2010; Raina et al., 2009, 2010) and included a suite of amino acids, carbohydrates, dimethylsulfonipropionate and filtered seawater (FSW) (Supplementary Information).
In situ chemotaxis assay (ISCA) experiments
Although the laboratory experiments were designed to provide a first glimpse into the chemotactic capacity of natural communities of coral-associated bacteria, laboratory-based measurements may be influenced by bottle effects, changes in community composition or the change in physical conditions from ocean to the laboratory. To examine coral–microbe chemotaxis within the natural coral reef environment, we complemented the laboratory experiments with in situ chemotaxis measurements, using a newly developed microfluidic-based platform, the ISCA. The ISCA was engineered using soft lithography techniques (Whitesides et al., 2001; Seymour et al., 2008) to create a high-throughput method for chemotaxis quantification in situ, allowing for the simultaneous and replicated testing of multiple chemoattractants under identical conditions, as well as on-chip controls. Each ISCA consists of a matrix of 24 cylindrical wells embedded within a ∼30-cm2 slab of the soft, inert polymer polydimethylsiloxane. Each well has a diameter of 10 mm and a height of 1 mm, resulting in a volume of ∼80 μl. Each well has two 1-mm diameter, 5-mm-high ports, which are openings connecting the interior of the chamber to the external environment. Individual wells are filled with 80 μl of putative chemoattractant using a pipette. Over the course of a 30-min deployment, the chemoattractant gradually leaks into the external environment through the two inlet ports via molecular diffusion, creating a gradient in the surrounding seawater that triggers the migration of chemotactic bacteria into the wells.
ISCA deployments were conducted 1 year subsequent to the laboratory experiments. The short branches of P. damicornis (used in the laboratory experiments) prevented the placement of ISCAs in between the coral branches, meaning that non-intrusive ISCA experiments close to the surface of this coral species were not possible. Consequently, we focussed this component of the study on the abundant coral species Acropora aspera and A. palifera, which have a more deeply branching structure that allowed for ISCAs to be placed in between coral branches (Supplementary Figure S2). ISCAs were deployed on the surface of A. aspera, located on Heron Island reef crest, and on the surface of A. palifera within lagoon (Supplementary Figure S1). These coral species represent the dominant species within these two regions of Heron Island reef (Wild et al., 2004). For each coral deployment, ISCAs were carefully inserted into the coral branches (Supplementary Figure S2), so that the wells were facing away from the centre of the coral. ISCAs were also deployed on the sandy substrate within Heron Island lagoon and within open water away from any coral (Supplementary Information; Supplementary Figure S1). We now describe the bacterial communities inhabiting each of these environments as ‘coral-associated’ and ‘non-coral-associated’ bacteria, respectively. For each water-type tested, two ISCAs were deployed in this fashion, so that samples could be collected for both 16S rRNA amplicon sequencing and flow cytometric analysis, respectively. For each ISCA deployment, four chemoattractants were tested simultaneously and each was replicated four times across the ISCA. These included: (1) an equimolar mix (100 μM each) of the amino acids arginine, histidine, isoleucine, leucine, lysine, methionine, phenylalanine, threonine, tryptophan and valine (Sigma-Aldrich, Sydney, NSW, Australia); (2) an equimolar mix (100 μM each) of the carbohydrates arabinose, fucose, galactose, glucose and mannose (Sigma-Aldrich); (3) 100 μM of ammonium chloride (Sigma-Aldrich); (4) 100 μM DMSP (Tokyo Chemical Industry, Tokyo, Japan). To avoid the generation of secondary chemical gradients, all chemoattractants for both syringe assays and ISCA deployments were diluted in 0.2 μm FSW, using seawater collected from the relevant sampling environment. This FSW was also used as a control in each experiment.
Chemotaxis sample preparation and analysis
For both the laboratory and in situ experiments, the intensity of chemotaxis was determined using flow cytometry to quantify the number of cells that migrated into syringes (laboratory experiments) and ISCA wells (in situ experiments). Upon completion of the assays, samples were immediately (within 10 min) fixed with glutaraldehyde (1% final concentration) for 20 min and frozen in liquid nitrogen before being stored at −80 °C. Samples were stained with SYBR Green I (1:10 000) (Invitrogen, Molecular Probes, Eugene, OR, USA) and analysed using a Becton Dickinson LSR II flow cytometer (BD Biosciences, San Jose, CA, USA). Bacterial populations were discriminated according to SYBR Green fluorescence and side-scatter (Marie et al., 1997; Seymour et al., 2007).
DNA extraction, 16S rRNA gene sequencing and analysis
DNA samples were quick-frozen in liquid nitrogen and stored at −80 °C. Prior to extraction, DNA samples were quick-thawed in a bath of hot water and a 5-μl aliquot from each sample was transferred into individual microfuge tubes. Genomic DNA was extracted from the samples using MicroLYSIS reagent (Microzone, Haywards Heath, UK) in a 1:5 dilution. The lysis protocol involved one cycle of the following 7-step thermocycling conditions: 65 °C for 15 min, 96 °C for 2 min, 65 °C for 4 min, 96 °C for 1 min, 65 °C for 1 min, 96 °C for 30 s and hold at 20 °C.
The four replicate samples of each chemoattractant across the ISCA designated for DNA collection were pooled to account for biological variability. The composition of the microbial communities responding to each chemoattractant was determined using 16S rRNA amplicon pyrosequencing. Extracted DNA was amplified using universal 16S primers 803F (5′-ATTAGATACCCTGGTAGTC-3′) and 1392R (5′-ACGGGCGGTGTGTRC-3′) (Supplementary Information). Amplicons were sequenced using the 454 GS-FLX pyrosequencing platform (Roche, Brandford, CT, USA). Homopolymer errors were removed using Acacia (Bragg et al., 2012), and 16S rRNA gene sequences were analysed using the QIIME pipeline (Caporaso et al., 2010; Kuczynski et al., 2011) (Supplementary Information).
Metagenomic analysis of bacterial communities
For each of the four water types where ISCAs were deployed, metagenomes from bulk seawater were also sequenced. For each water type, 10 l of seawater was collected in sterile 10-l Schott bottles (Supplementary Information). Shotgun metagenomic libraries were generated for each of the four samples using the 454 pyrosequencing platform (454 GS-FLX, Roche). Further details of metagenomic sequencing, data quality control and analysis are provided in Tout et al. (2014). For this study, post quality control sequence analysis focussed on genes associated with bacterial chemotaxis and motility, which were identified by comparing sequences to the KEGG database at the hierarchal functional level one (Supplementary Information).
Chemotactic index and statistical analysis of data
For both laboratory and in situ experiments, the accumulation of bacteria in response to the chemoattractants was expressed in terms of a chemotactic index, Ic (mean±s.d.). The Ic value was calculated by normalising the concentration of cells responding to a given chemoattractant to the number of cells responding to the corresponding FSW control. Positive chemotaxis was identified when Ic was significantly (P<0.05) greater than 1. Ic data were tested for normality using the Kolmogorov–Smirnov test, and Levene’s test was used to test for homogeneity of variance. In cases where these assumptions were not met, log10 transformations were performed. To compare chemotaxis levels in the laboratory experiments, three-way analysis of variance was used (water type × chemoattractant × concentration). For the ISCA experiments, chemotactic responses within water types were compared using a paired T-test (chemoattractant), whereas a two-way analysis of variance was used to determine differences between the water types (water type × chemoattractant).
A network analysis approach was employed to examine chemotactic preferences at the level of the OTUs (Operational Taxonomic Units). This provided information on whether chemotactic OTUs demonstrated a specialist response, whereby they were only chemotactic to a specific chemoattractant, or a generalist response, whereby they exhibited chemotaxis towards multiple attractants. OTUs responding to the tested chemoattractants were plotted using Cytoscape 2.8.3 (www.cytoscape.org) (Shannon et al., 2003; Smoot et al., 2011; Fuhrman and Steele, 2008; Fan et al., 2012; Needham et al., 2013) according to the QIIME pipeline using the force-directed Cytoscape layout: edge-weighted spring embedded according to e-weights (Kamada and Kawai, 1989; Caporaso et al., 2010; Kuczynski et al., 2011) (Supplementary Information). Known coral-associated OTUs where identified according to Raina et al. (2010) and were colour coded to highlight their distribution among the chemoattractants.
Differences in the relative abundance of motility and chemotaxis genes between the four metagenomes were identified using Fisher’s exact test with a Benjamini false discovery rate multiple test correction, within the statistical analysis of metagenomic profiles package (Parks and Beiko, 2010). All quoted q-values represent corrected values, with only values of <0.05 reported (Parks and Beiko, 2010). Confidence intervals (95%) were determined using the Newcombe–Wilson method. Multivariate statistical software (PRIMER v6) was used to measure the degree of similarity between metagenomes (Clarke and Gorley, 2006). Data were square-root transformed and the Bray–Curtis similarity was calculated between samples. SIMPER analysis (Clarke, 1993) was used to identify the metabolic categories contributing most to the dissimilarity between the metagenomes.
Results
Laboratory chemotaxis experiments
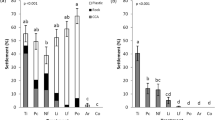
DMSP invoked a strong chemotactic response by bacteria from coral-associated water, as evidenced by cell concentrations in DMSP-containing syringes reaching 2–15 times higher values than the control (that is, Ic=2–15; Figure 1a). However, no significant difference was measured in the total number of cells responding to different concentrations of DMSP (Figure 1a; Supplementary Table S1). Coral-associated bacteria exhibited significantly (P<0.05) higher levels of chemotaxis towards all concentrations of DMSP (Ic=2–15) than the non-coral-associated bacteria, which did not display chemotaxis towards any of the tested DMSP concentrations (Ic=0.11–0.18) (Figures 1a and b; Supplementary Table S1).
Chemotactic index, Ic, of natural bacterial assemblages responding to 10 μM, 100 μM and 1 mM concentrations of DMSP (a and b), amino acids (c and d) and carbohydrates (e and f) and the FSW control (line at 1 on the y axis). Bacterial concentrations in syringes containing chemoattractants were normalised to concentrations in the FSW control. Responses above the horizontal line represent positive chemotaxis. (a, c and e) Coral-reef-associated bacteria. (b, d and f) non-coral-associated bacteria. Vertical bars represent mean±s.d. (n=3). An asterisk on the x axis represents chemotaxis that is significantly higher than the FSW control; α=0.05. C represents the concentration where a significantly highest chemotactic response occurred; α=0.05.
Among the coral-associated bacteria collected from seawater adjacent to P. damicornis, significant (P<0.001) positive chemotaxis was also observed towards several of the tested amino acids. The strongest chemotactic response in the coral-associated bacteria was to 1 mM tryptophan (Ic=28.1±4.9 s.d.) (Figure 1c), where Ic increased with higher concentrations of the attractant and chemotaxis to all concentrations of tryptophan was significantly greater than to the FSW control. Significant (P<0.001) chemotaxis towards all concentrations of aspartic acid (Ic=5.2±3.4 s.d.) and casamino acid (Ic=15.0±13.6 s.d.) was also measured (Figure 1c; Supplementary Table S2), where the 10-μM concentration invoked the highest response in both cases. Responses varied significantly according to concentration, but across all of the tested amino acids there was no general trend of increasing or decreasing Ic with increasing amino-acid concentration. Bacteria from the coral-associated seawater exhibited significantly (P<0.001) higher levels of chemotaxis towards aspartic acid, tryptophan and casamino acids than the non-coral-associated bacteria (Figures 1c and d; Supplementary Table S2). The non-coral-associated bacteria also showed chemotaxis towards some amino acids, but the levels of chemotaxis were significantly lower (P<0.001). The most potent chemoattractants for these non-coral-associated bacteria also differed (P<0.001) from the coral-associated bacteria and included valine, lysine and threonine (Figure 1d).
None of the tested carbohydrates induced a significant chemotactic response by coral-associated bacteria (Figure 1e; Supplementary Table S3), whereas non-coral-associated bacteria showed positive chemotaxis towards all concentrations of arabinose (P=0.001), with a maximum response (Ic=5.8±0.7 s.d.) at a concentration of 100 μM (Figure 1f; Supplementary Table S3).
ISCA experiments
For each of the tested chemoattractants, the chemotactic responses were significantly greater in the ISCAs deployed on the surface of A. aspera in the reef crest (P<0.001) (Figure 2; Supplementary Table S4) than at any other water type. For all other tested water types, there was no positive chemotaxis to any chemoattractant (Figure 2). However, for the ISCAs deployed on the surface of A. aspera in the reef crest, there was significant chemotaxis towards DMSP (Ic=3.2±0.6 s.d., P<0.05, T=6.18, Supplementary Table S5), the amino-acid mixture (Ic=25.1±7.9 s.d., P<0.05, T=7.50, Supplementary Table S6) and ammonium chloride (Ic=3.5±0.5 s.d., P<0.05, T=7.97, Supplementary Table S7) (Figure 2). There was a response to the carbohydrate mixture (Ic=8.7±4.3 s.d.), but due to high inter-replicate variability this was not significantly different from the FSW control (Supplementary Table S8).
Bacterial chemotaxis to the amino-acid mix, carbohydrate mix, DMSP and ammonium chloride (all at 100 μM) observed at four coral reef microenvironments on Heron Island using the in situ chemotaxis assay (ISCA). ISCAs were deployed in the open water outside the reef (blue bars); on the coral A. aspera on the reef crest (red bars); on the coral A. palifera in the lagoon (green bars); and at the sandy substrate under water in the lagoon (orange bars) (see Supplementary Figure S1). Values on the y axis represent the chemotactic index, Ic, computed after bacterial concentrations in ISCA wells containing chemoattractants were normalised to bacterial concentrations in the FSW control. Responses above the horizontal line represent positive chemotaxis. Vertical bars represent mean±s.d. (n=4). An asterisk above a microenvironment represents chemotaxis that is significantly higher than the FSW control; α=0.05. A full colour version of this figure is available at the International Society for Microbial Ecology journal online.
Community composition of chemotactic bacteria from the ISCAs
The composition of the chemotactic bacteria in the A. aspera ISCA deployments conducted on the reef crest (Figure 3) differed from the community composition in the surrounding seawater (Supplementary Figure S3). The background seawater sample was dominated by microorganisms that are typically the most abundant members of bacterial communities within oligotrophic tropical waters, including Candidatus pelagibacter (SAR11) and Synechococcus (Supplementary Figure S3).
Taxonomic identities (family) of chemotactic bacteria responding to the ISCA deployments on the reef-crest coral A. aspera using 16S rRNA gene sequencing, where hits were generated by comparing the sequences with BLASTn to the Greengenes database in QIIME. The relative proportions of families responding to each chemoattractant were normalised by multiplying by the cell abundance retrieved from FCM (Figure 1) (Dennis et al., 2013). The microbial community identified in the FSW control is representative of organisms that swam into this treatment as a consequence of random motility, rather than chemotaxis, due to the lack of any chemical gradient. As such, this sample provides an overview of the motile, but not necessarily chemotactic proportion of the community. For more detail on unique taxa responding to individual chemoattractants, see Supplementary Figure S3.
At the family level, the community retrieved from the ISCA wells were relatively conserved across the chemoattractants tested and was dominated by members of the Rhodobacteraceae, Comamonadaceae, Flavobacteriaceae, Pseudomonadaceae and Sphingomonadaceae (Figure 3; Supplementary Figure S4a). However, some less-abundant families displayed a more specialized response, with representatives responding to only one or a subset of the tested chemoattractants (Supplementary Figure S4b). For example, Vibrionaceae responded only to amino acids, carbohydrates and DMSP, whereas the Shewanellaceae responded only to DMSP and carbohydrates (Figure 3).
The extent of ‘generalism’ versus ‘specialism’ in the chemotactic responses to individual chemoattractants was probed more extensively at the OTU level using network analysis (Figure 4). Of the 394 OTUs responding to the tested chemoattractants, only 4.3% responded to all four compounds (the nodes in the centre of the network in Figure 4) and can be considered ‘generalist chemotaxers’. Of this ‘generalist’ community, 65% were identified as belonging to known coral-associated families. On the other hand, 19.3% of OTUs responded to two or three chemoattractants, of which 44% were from known coral-associated families. The bulk of OTUs (76.4%) exhibited a ‘specialist’ response, being attracted to only one compound, 39.5% of which belonged to known coral-associated families. Of these, 42.5% responded to the carbohydrate mixture only, 24.9% to the amino-acid mixture only, 16.6% to DMSP only and 15.9% to ammonium chloride only. These specialist OTUs included a diverse range of organisms often belonging to families that contain known coral associates, including Endozoicomonaceae, Alteromonadaceae and Vibrionaceae (Figure 4).
OTU network resulting from the ISCA deployment on A. aspera. Chemotactic bacterial OTUs (small circles) are linked to each chemoattractant that they responded to (coloured diamonds) by lines (edges) with colours corresponding to each chemoattractant. Where an OTU responded only to a single chemoattractant, its corresponding node is connected only to that chemoattractant (the outer clusters of nodes). Where an OTU responded to multiple chemoattractants, the corresponding node is connected to the relevant groups of chemoattractants (nodes in the centre). OTUs belonging to known groups of coral-associated bacteria, according to Raina et al. (2010), are denoted by the different colours listed in the legend. These known coral associates account for 41.37% of all OTUs. Of the remaining black nodes, ∼27.9% of OTUs could not be identified beyond the order level. A full colour version of this figure is available at the International Society for Microbial Ecology journal online.
Chemotaxis gene patterns in metagenomes
Patterns in chemotaxis and motility genes obtained from metagenomic data closely reflect the phenotypic patterns observed in each of the chemotaxis experiments. Genes encoding cell motility and chemotaxis, including the chemotaxis genes cheA, B, D, R, V, W, Y, Z, the methyl-accepting chemotaxis gene MCP, the motility genes motA, B, C and the flagellar proteins flgA, B, D, E, G, H, I, L, M, flhA, B, C, D and fliC, F, G, I, M, N, Y, O, Z, P, R, were responsible for driving significant differences between the coral-associated and the non-coral-associated metagenomes (Supplementary Tables S9–S11). These motility genes were over-represented in the coral-associated water types, comprising 0.83% of the total number of sequences (Table 1), compared with only 0.23% in the open-water type (Figure 5a). Consistent with the significantly higher levels of chemotaxis observed in A. aspera in the reef crest, the above mentioned genes were also generally significantly (q<0.05, Fisher’s exact test) over-represented on the surface of A. aspera (Figures 5a and b). In A. aspera, MCP, cheA, motA, motB, cheB, cheR and cheY were significantly (q<0.05) over-represented relative to the open-water type. The differences between the two coral-associated samples were less pronounced: MCP was the only gene significantly over-represented in A. aspera on the reef crest relative to A. palifera in the lagoon (q<0.05) (Figure 5b).
Functional categories of motility and chemotaxis composition of four metagenomic libraries derived from different reef habitats on Heron Island by comparing the sequences with BLASTn to the KEGG database. (a) Cell motility category, and (b) functional genes associated with the cell motility category. Relative representation in the metagenome was calculated by dividing the number of hits to each category by the total number of hits to all categories, thus normalizing by sequencing effort.
Discussion
Our data provide the first direct in situ measurements of chemotactic behaviour among natural populations of coral-associated bacteria, with high levels of chemotaxis demonstrated towards a range of coral holobiont-related chemicals. These in situ measurements add a new dimension to existing evidence that some coral pathogens are chemotactic towards coral mucus and other host-derived compounds (Banin et al., 2001; Koren and Rosenberg, 2006; Rosenberg et al., 2007; Meron et al., 2009; Kimes et al., 2011; Vidal-Dupiol et al., 2011; Garren et al., 2014), and that chemotaxis and motility genes can be prominent and dynamic features in the metagenomes of coral–microbial communities (Vega Thurber et al., 2009). Given the strong chemotactic responses that we observed in the laboratory experiments and ISCA deployments, we propose that motility and chemotaxis may be important phenotypes within the context of coral–microbial interactions.
Our laboratory-based and in situ measurements indicate that coral reef bacteria are well equipped to sense and respond to the chemicals released from corals. The diffusion of chemicals away from the coral surface into the surrounding seawater generates microscale chemical gradients extending into the adjacent seawater, whereas the shedding of carbon-rich mucus by corals may produce chemical hotspots (Wild et al., 2004). Chemotaxis is thus likely to be an important mechanism favouring the colonisation of coral surfaces by bacteria.
The laboratory-based syringe assays revealed a much stronger chemotactic response by coral-associated bacteria, obtained from near the surface of the coral species P. damicornis, than by bacteria collected from a non-coral site. In addition, the bacteria from the coral- and non-coral-associated sites showed different preferences for chemoattractants, indicating that the chemical requirements, or use of chemical signals as chemotactic cues, vary between coral-associated and nearby open-water bacterial communities. These patterns were supported by the results of the in situ experiments, whereby bacteria associated with the surface of the coral A. aspera exhibited significantly higher levels of chemotactic capability than bacteria from any other water type around the reef or from the open water.
The short-branching morphology of P. damicornis was not amenable to ISCA deployment, so ISCA experiments were performed on the more deeply branching A. aspera in the reef crest and A. palifera in the lagoon. Although this prevents direct comparison of our laboratory and in situ results, the general pattern of higher levels of chemotaxis on coral surfaces was conserved between approaches.
Elevated levels of chemotaxis observed among the microbial communities associated with the reef crest coral A. aspera in the ISCA experiments were supported by metagenomic analysis, where genes associated with motility and chemotaxis were significantly more abundant in the seawater associated with A. aspera than in any other sample. Of note, motility and chemotaxis genes in the A. aspera sample were significantly more abundant than in the sample obtained from the other coral species A. palifera located in the lagoon, which is directly in line with the differences in chemotactic responses observed between the bacterial communities associated with these two coral species in the ISCA experiments. Taken together, our results demonstrate that chemotaxis is heterogeneous across a coral reef and between communities associated with different corals. The different chemtoaxis patterns observed between the bacterial communities associated with the two Acropora species may be due to inter-coral variability in microbial community composition and function (Rohwer et al., 2001, 2002; Tout et al., 2014), or slight differences in the morphology of the two coral species may have altered the biophysical environment inhabited by the resident microbes (Wallace, 1999). In addition, the location of the corals within the Heron Island reef system may be responsible for some of these differences. A. aspera was chosen for sampling in the reef crest environment, whereas A. palifera was chosen within the lagoon, because these species dominate the coral communities within these two habitats, respectively (Wild et al., 2004). The reef crest, where the A. aspera deployment occurred is an environment characterised by a wall of Acropora corals, where 69% of the benthos is covered by hard corals (Salmond et al., 2013). In contrast, in the lagoon where the ISCAs were deployed on A. palifera, hard corals contribute to only 6% of the total benthic cover (the lowest hard coral cover on Heron Island) (Salmond et al., 2013) and the bulk of the substrate is sand. As a consequence, the local environment surrounding the different coral colonies varied substantially, with differences in the bulk microbial community within these two habitats potentially contributing to the differences observed here. Finally, Vega Thurber et al. (2009) showed significant shifts in chemotaxis genes among the microbial communities associated with stressed corals relative to healthy individuals, and although no notable signs of disease or stress were apparent in either coral tested here, there remains the possibility that differences in the health status of the coral hosts may contribute to differences in the level of chemotaxis observed here.
Natural communities of coral-associated bacteria showed strong chemotactic responses towards DMSP in both the laboratory assays and the ISCA deployments. This is consistent with DMSP being an important source of carbon and reduced sulphur for marine bacteria (Howard et al., 2006) and a chemical cue for some coral pathogens (Garren et al., 2014). DMSP is abundant on coral reefs (Broadbent et al., 2002; Broadbent and Jones, 2004; Raina et al., 2010), with coral mucus concentrations (up to 62 μM) the highest reported in the marine environment (Broadbent et al., 2002; Broadbent and Jones, 2004). There is now evidence that both the coral symbionts Symbiodinium spp. (Keller et al., 1989; Broadbent et al., 2002) and the coral animal (Raina et al., 2013) have the capacity to produce significant quantities of DMSP. Our results confirm that, similar to other marine microorganisms (Miller et al., 2004; Seymour et al., 2010; Sharp et al., 2012), coral-reef-associated bacteria use chemotaxis to enhance their access to DMSP or to follow DMSP gradients as a cue to locate the host (Garren et al., 2014). The dominant bacterial taxa responding to DMSP included known coral-associated species that have the capacity to degrade DMSP (Flavobacteriaceae) and DMS (Comamonadaceae) or both (Rhodobacteracea, Pseudomonaceae and Halomonaceae) (Raina et al., 2010). It is notable that we also observed some Vibrio sequences in the DMSP sample, because recent evidence suggests that DMSP can be a potent chemoattractant for coral pathogens belonging to this genus (Garren et al., 2014).
Strong chemotactic responses to several amino acids by coral-associated bacteria were also observed using both the syringe assays and the ISCA deployments. The amino acids used in the chemotaxis assays have previously been found in the mucus of several coral species (Ducklow and Mitchell, 1979b; Meikle et al., 1988; Fitzgerald and Szmant, 1997). Chemotaxis towards amino acids is well recognised among enteric bacteria (Adler, 1966; Mesibov and Adler, 1972; Kim et al., 2001; Bainer et al., 2003; Frank et al., 2011), has been demonstrated in some marine bacteria (Barbara and Mitchell, 2003a, 2003b) and our results indicate that it may also be involved in coral–microbe interactions in coral reef environments.
Marine bacteria have been shown to exhibit chemotaxis towards carbohydrates present in the exudates of phytoplankton (Hellebust, 1965; Bell and Mitchell, 1972) and in the mucus of the Hawaiian squid Euprymna scolopes light organ (DeLoney-Marino et al., 2003). Despite the prominence of carbohydrates in coral mucus (Ducklow and Mitchell, 1979b; Meikle et al., 1988; Wild et al., 2005), the chemotactic response of coral-associated bacteria towards the tested carbohydrates in both the syringe assays and the ISCAs were never significantly different from the FSW control. Taken together, these findings indicate that carbohydrates previously shown to occur in coral mucus do not appear to be an important chemical cue for coral-associated bacteria.
Our results demonstrate that natural populations of coral-associated bacteria exhibit chemotaxis towards ammonium. Aquatic heterotrophic bacteria have previously been shown to use chemotaxis to exploit patches of inorganic substrates including ammonium (Stocker and Seymour, 2012; Dennis et al., 2013). Within coral reefs, nitrogen is often a limiting nutrient in the water column (Thomas and Owen, 1971; Crossland et al., 1984; Moore et al., 2013), as nitrate and ammonium concentrations are often <2 μM (Crossland et al., 1984; Bythell, 1990). However, ammonium and other inorganic nutrients are likely to be generated as metabolic by-products in the coral holobiont (Kawaguti, 1953; Muscatine and D’Elia, 1978; Siboni et al., 2008), and ammonium levels can reach up to 50 μM within coral mucus (Wild et al., 2005). Therefore the ability to use chemotaxis to exploit elevated levels of inorganic nutrients near the surface of corals may provide a competitive advantage for some coral reef bacteria.
The microbial community identified in the FSW control is representative of organisms that swam into this treatment as a consequence of random motility, rather than chemotaxis, due to the lack of any chemical gradient. As such, this sample provides an overview of the motile, but not necessarily chemotactic proportion of the community. The choice of chemoattractant concentrations in our experiments was guided by concentrations previously shown to occur within coral microenvironments, such as coral mucus (Broadbent and Jones, 2004; Wild et al., 2004, 2005, 2010). In most cases, no significant difference was observed in the chemotactic response between different concentrations. However, the coral-associated bacteria’s response to tryptophan increased with increasing concentrations. On the other hand, in the case of aspartic and casamino acids, the strongest chemotaxis occurred in response to the lowest tested concentration of 10 μM, which is perhaps indicative of an inhibitory response associated with saturation of chemoreceptors at higher concentrations (Mesibov et al., 1973). These patterns indicate that the chemotactic sensitivities and thresholds for coral-associated bacteria vary between different types of compounds, which may reflect the relative availability and concentrations of these substrates in the environment.
Recent measurements of the chemotactic behaviour of a natural bacterial population from a lake revealed a strong phylogenetic partitioning in the response towards inorganic compounds (Dennis et al., 2013). Our results have expanded upon this work by identifying members of the chemotactic bacterial communities associated with the coral species A. aspera, and demonstrating that these differed substantially from the community in the surrounding seawater. This indicates that chemotaxis may act as a behavioural filter, favouring certain species over others in associating with corals and thus determining the composition of microbial communities within specific coral microniches.
At the family level of taxonomic resolution, we found that the same bacterial groups were typically the dominant responders to all chemoattractants, indicating that microorganisms belonging to a restricted range of bacterial families perform chemotaxis in the coral reef environment. These groups included families with members that have metabolic requirements for the tested chemoattractants (for example, Flavobacteriaceae for DMSP; Raina et al., 2010) or are known coral associates (Rhodobacteraceae, Comamonadaceae, Flavobacteriaceae and Pseudomonadaceae (Raina et al., 2010; Morrow et al., 2012)). However, at a finer taxonomic resolution it became clear that specialist chemotactic responses, whereby organisms only exhibited chemotaxis to certain chemicals, were common. There were some families where representatives only responded to one or a subset of the chemoattractants tested. These more-specialised groups of chemotaxers included members of the Shewanellaceae and Vibrionaceae families. The Shewanellaceae only exhibited chemotaxis towards DMSP and carbohydrates. This family includes known coral-associated bacteria (Shnit-Orland and Kushmaro, 2009; Raina et al., 2010; Ceh et al., 2012) and species that degrade DMSP (Raina et al., 2010). Chemotaxis by members of the marine Shewanellaceae towards amino acids and algae has previously been observed (Barbara and Mitchell, 2003a, 2003b), and our results demonstrate that members of this known group of coral associates may use chemotaxis towards DMSP within the coral holobiont.
Clear differentiation of the chemotactic community into groups of ‘specialist’ and ‘generalist’ chemotaxers became particularly evident when responses were assessed at the OTU level. Approximately 4.3% of all OTUs responded to all four compounds tested, whereas 19.3% responded to 2 and 3 chemoattractants. This suggests that a subset of the chemotactic community is made of ‘generalist chemotaxers’ that have the capacity to sense and direct movement in response to diverse chemicals compounds. Currently, little is known about the chemoreceptors used by marine bacteria (Stocker and Seymour, 2012), but the generalist chemotaxers identified here may either have multiple sets of chemoreceptors, which allow them to respond to a variety of compounds (Wadhams and Armitage, 2004; Parales et al., 2013), or single chemoreceptors that allow for binding of multiple compounds (Adler, 1969; Glekas et al., 2012). However, the bulk (76.4%) of chemotactic OTUs identified here can be classified as ‘specialist chemotaxers’, as they responded to only one compound. Many of these specialist chemotaxers were also known coral associates including members of Endozoicomonaceae, Rhodobacteraceae, Vibrionaceae and Mycobacteriacea. In the environment, the heterogeneity in chemotactic responses observed here could lead to strong niche partitioning among the bacterial community, suggesting that this behaviour could underpin some of the heterogeneity in microbial community composition previously observed between different coral hosts (Rohwer et al., 2002).
The partitioning of coral–microbe communities observed here may be encouraged by differential release of chemoattractants from the coral holobiont under varying environmental conditions. There is evidence that the production and release of key chemoattractants, such as DMSP, can vary markedly under different environmental conditions and coral health states (Raina et al., 2013; Garren et al., 2014). From a bacterium’s perspective, chemotaxis serves a variety of potential ecological functions, including (i) providing cells with greater access to important growth substrates and nutrients in otherwise oligotrophic habitats (Stocker et al., 2008; Seymour et al., 2009, ii) enhancing exposure to terminal electron acceptors/donors (Schweinitzer and Josenhans, 2010) or (iii) use of infochemicals that may provide pathogenic microorganisms with information about the location, health and potential susceptibility of coral hosts to infection (Garren et al., 2014). Heterogeneity in the strength of chemotaxis towards different chemoattractants observed across the different water types is perhaps indicative of varying ecological strategies among microbial communities on coral reefs.
Conclusions
We have provided the first in situ quantification of bacterial chemotaxis on a coral reef. Previous studies have shown that different habitats and microenviroments on coral reefs host phylogenetically distinct communities (Rohwer et al., 2002; Rohwer and Kelly, 2004), but our results show that different reef features (for example, coral species and water types) also host distinct microbial phenotypic capacities, specifically the ability to perform chemotaxis. In addition, we have provided a direct mechanism for how chemical gradients associated with coral surfaces may be involved in the establishment of specific coral–bacterial relationships, and how microbial chemotaxis might shape the composition of coral reef bacterial communities. As such, we suggest that within the chemically and physically complex coral microenvironment, bacterial behaviours, including motility and chemotaxis, may be fundamental drivers of patterns in microbial diversity and metabolism, coral infection dynamics and chemical cycling processes.
References
Adler J . (1966). Chemotaxis in bacteria. Science 153: 708–716.
Adler J . (1969). Chemoreceptors in bacteria. Science 166: 1588–1597.
Adler J . (1973). A method for measuring chemotaxis and use of the method to determine optimum conditions for chemotaxis by Escherichia coli. J Gen Microbiol 74: 77–91.
Bainer R, Park H, Cluzel P . (2003). A high-throughput capillary assay for bacterial chemotaxis. J Microbiol Methods 55: 315–319.
Banin E, Israely T, Fine M, Loya Y, Rosenberg E . (2001). Role of endosymbiotic zooxanthellae and coral mucus in the adhesion of the coral-bleaching pathogen Vibrio shiloi to its host. FEMS Microbiol Lett 199: 33–37.
Barbara GM, Mitchell JG . (2003a). Marine bacterial organisation around point-like sources of amino acids. FEMS Microbiol Ecol 43: 99–109.
Barbara GM, Mitchell JG . (2003b). Bacterial tracking of motile algae. FEMS Microbiol Ecol 44: 79–87.
Bartlett DG, Matsumura P . (1986). Behavioural responses to chemical cues by bacteria. J Chem Ecol 12: 1071–1089.
Bell W, Mitchell R . (1972). Chemotactic and growth responses of marine bacteria to algal extracellular products. The Biol Bull 143: 265–277.
Bianchi A, Giuliano L . (1996). Enumeration of viable bacteria in the marine pelagic environment. Appl Microbiol 62: 174–177.
Blackburn N, Fenchel T, Mitchell J . (1998). Microscale nutrient patches in planktonic habitats shown by chemotactic bacteria. Science 282: 2254–2256.
Bragg L, Stone G, Imelfort M, Hugenholtz P, Tyson GW . (2012). Fast, accurate error-correction of amplicon pyrosequences using Acacia. Nat Methods 9: 425–426.
Broadbent AD, Jones GB, Jones RJ . (2002). DMSP in corals and benthic algae from the Great Barrier Reef. Estuar Coast Shelf Sci 55: 547–555.
Broadbent AD, Jones GB . (2004). DMS and DMSP in mucus ropes, coral mucus, surface films and sediment pore waters from coral reefs in the Great Barrier Reef. Mar Freshwater Res 55: 849–855.
Bythell JC . (1990). Nutrient uptake in the reef-building coral Acropora palmate at natural environmental concentrations. Mar Ecol Prog Ser 68: 65–69.
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336.
Ceh J, van Keulen M, Bourne DG . (2011). Coral-associated bacterial communities on Ningaloo Reef, Western Australia. FEMS Microbiol Ecol 75: 134–144.
Ceh J, Raina J-B, Soo RM, van Keulen M, Bourne DG . (2012). Coral-bacterial communities before and after a coral mass spawning event on Ningaloo Reef. PLoS One 7: e36920.
Chet I, Mitchell R . (1976). Ecological aspects of microbial chemotactic behaviour. Annu Rev Microbiol 30: 221–239.
Clarke KR, Gorley RN . (2006) PRIMER v6: User Manual/Tutorial. PRIMER-E: Plymouth, UK.
Clarke KR . (1993). Non-parametric multivariate analyses of changes in community structure. Aust J Ecol 18: 117–143.
Crossland CJ, Hatcher BG, Atkinson MJ, Smith SV . (1984). Dissolved nutrients of a high-latitude coral reef, Houtman Abrolhos Islands, Western Australia. Mar Ecol Prog Ser 14: 159–163.
DeLoney-Marino CR, Wolfe AJ, Visick KL . (2003). Chemoattraction of Vibrio fischeri to Serine, nucleosides and N-Acetylneuraminic Acid, a component of squid light-organ Mucus. Appl Environ Microbiol 69: 7527–7530.
Dennis PG, Seymour J, Kumbun K, Tyson GW . (2013). Diverse populations of lake water bacteria exhibit chemotaxis towards inorganic nutrients. ISME J 7: 1661–1664.
Ducklow HW, Mitchell R . (1979a). Bacterial populations and adaptations in the mucus layers of living corals. Limnol Oceanogr 24: 715–725.
Ducklow HW, Mitchell R . (1979b). Composition of mucus released by coral reef coelenterates. Limnol Oceanogr 24: 706–714.
Fan L, Reynolds D, Liu M, Stark S, Kjelleberg S, Webster NS, Thomas T . (2012). Functional equivalence and evolutionary convergence in complex communities of microbial sponge symbionts. Proc Natl Acad Sci USA 109: E1878–E1887.
Fitzgerald LM, Szmant AM . (1997). Biosynthesis of ‘essential’ amino acids by scleractinian corals. Biochem J 322: 213–221.
Frank V, Koler M, Furst S, Vaknin A . (2011). The physical and functional thermal sensitivity of bacterial chemoreceptors. J Mol Biol 411: 554–566.
Frias-Lopez J, Zerkl AL, Bonheyo GT, Fouke BW . (2002). Partitioning of bacterial communities between seawater and healthy, black band diseased and dead coral surfaces. Appl Environ Microbiol 68: 2214–2228.
Fuhrman J, Steele JA . (2008). Community structure of marine bacterioplankton: patterns, networks, and relationships to function. Aquat Microb Ecol 53: 69–81.
Garren M, Son K, Raina JB, Rusconi R, Menolascina F, Shapiro OH et al. (2014). A bacterial pathogen uses dimethylsulfoniopropionate as a cue to target heat-stressed corals. ISME J 8: 999–1007.
Glekas GD, Mulhern BJ, Kroc A, Duelfer KA, Lei V, Rao CV, Ordal GW . (2012). The Bacillus subtilis chemoreceptor McpC senses multiple ligands using two discrete mechanisms. J Biol Chem 287: 39412–39418.
Grossart HP, Riemann L, Azam F . (2001). Bacterial motility in the seas and its ecological implications. Aquat Microb Ecol 25: 247–258.
Hellebust JA . (1965). Excretion of some organic compounds by marine phytoplankton. Limnol Oceanogr 10: 192–206.
Hill RW, Dacey WH, Krupp DA . (1995). Dimethylsulfonioprpionate in reef corals. Bull Mar Sci 57: 489–494.
Hoppe HG . (1976). Determination and properties of actively metabolising heterotrophic bacteria in the sea, investigated by means of micro-autoradiography. Mar Biol 36: 291–302.
Howard EC, Henriksen JR, Buchan A, Reisch CR, Burgmann H, Welsh R et al. (2006). Bacterial taxa that limit sulfur flux from the ocean. Science 314: 649–652.
Jannasch HW, Jones GE . (1959). Bacterial populations in seawater as determined by different methods of enumeration. Limnol Oceanogr 4: 128–139.
Kamada T, Kawai S . (1989). An algorithm for drawing general undirected graphs. Inform Process Lett 31: 7–15.
Kawaguti S . (1953). Ammonium metabolism of reef corals. Biol J Okayama Univ 1: 171–176.
Keller MD, Bellows WK, Guillard RL . (1989). Dimethylsulfide production in marine phytoplankton. In Saltzman ES, Cooper JC (eds) Biogenic Sulfur in the Environment. American Chemical Society: Washington, DC, pp 167–182.
Kellogg C . (2004). Tropical Archaea: diversity associated with the surface microlayers of corals. Mar Ecol Prog Ser 273: 81–88.
Kim C, Jackson M, Lux R, Khan S . (2001). Determinants of chemotactic signal amplification in Escherichia coli. J Mol Biol 307: 119–135.
Kimes NE, Grim CJ, Johnson WR, Hasan NA, Tall BD, Kothary MH et al. (2011). Temperature regulation of virulence factors in the pathogen Vibrio coralliilyticus. ISME J 6: 835–846.
Koren O, Rosenberg E . (2006). Bacteria associated with mucus and tissues of the coral Oculina patagonica in summer and winter. Appl Environ Microbiol 728: 5254–5259.
Kuczynski J, Stombaugh J, Walters WA, Gonzalex A, Caporaso JG, Knight R . (2011). Using QIIME to analyse 16S Rrna gene sequences from microbial communities. Curr Protoc Bioinformatics Chapter 10: Unit 10.7.
Kuhl M, Cohen Y, Dalsgaard T, Jørgensen BB, Revsbech NP . (1995). Microenvironment and photosynthesis of zooxanthellae in sceractinian corals studied with microsensors for O2, pH, and light. Mar Ecol Prog Ser 117: 159–172.
Marie D, Partensky F, Jacquet S . (1997). Enumeration and cell cycle analysis of natural populations of marine picoplankton by flow cytometry using the nucleic acid stain SYBR Green I. Appl Environ Microbiol 63: 186–193.
Mass T, Genin A, Shavit U, Grinstein M, Tchernov D . (2010). Flow enhances photosynthesis in marine benthic autotrophs by increasing the efflux of oxygen from the organism to the water. Proc Natl Acad Sci USA 107: 2527–2531.
Meikle P, Richards GN, Yellowlees D . (1988). Structural investigations on the mucus from six species of coral. Mar Biol 99: 187–193.
Meron D, Efrony R, Johnson WR, Schaefer AL, Morris PJ, Rosenberg E et al. (2009). Role of flagella in virulence of the coral pathogen Vibrio coralliilyticus. Appl Environ Microbiol 75: 5704–5707.
Mesibov R, Adler J . (1972). Chemotaxis toward amino acids in Escherichia coli. J Bacteriol 112: 315–326.
Mesibov R, Ordal GW, Adler J . (1973). The range of attractant concentrations for bacterial chemotaxis and the threshold and size of response over the range. J Gen Physiol 62: 203–223.
Miller TR, Hnilicka K, Dziedzic A, Desplats P, Belas R . (2004). Chemotaxis of Silicibacter sp. strain TM1040 toward dinoflagellate products. Appl Environ Microbiol 70: 4692–4701.
Mitchell JG, Pearson L, Bonazinga A, Dillon S, Khoury H, Paxinos R . (1995). Long lag times and high velocities in the motility of natural assemblages of marine bacteria. Appl Environ Microbiol 61: 877–882.
Mitchell JG, Pearson L, Dillon S . (1996). Clustering of marine bacteria in seawater enrichments. Appl Environ Microbiol 62: 3716–3721.
Moore CM, Mills MM, Arrigo KR, Berman-Frank I, Bopp L, Boyd PW et al. (2013). Processes and patterns of oceanic nutrient limitation. Nat Geosci 6: 701–710.
Morrow KM, Moss AG, Chadwick NE, Liles MR . (2012). Bacterial associates of two Caribbean coral species reveal species-specific distribution and geographic variability. Appl and Environ Microbiol 78: 6438–6449.
Muscatine L, Cernichiari E . (1969). Assimilation of photosynthetic products of Zooxanthellae by a reef coral. Biol Bull 137: 506–523.
Muscatine L, D’Elia CFD . (1978). The uptake, retention and release of ammonium by reef corals. Limnol Oceanogr 23: 725–734.
Needham DM, Chow CET, Cram JA, Sachdeva R, Parada A, Fuhrman JA . (2013). Short-term observation of marine bacterial and viral communities: patterns, connections and resilience. ISME J 7: 1274–1285.
Parales RE, Luu RA, Chen GY, Liu X, Wu V, Lin P et al. (2013). Pseudomonas putida F1 has multiple chemoreceptors with overlapping specificity for organic acids. Microbiology 159: 1086–1096.
Parks DH, Beiko RG . (2010). Identifying biologically relevant differences between metagenomic communities. Bioinformatics 26: 715–721.
Paul JH, DeFlaun MF, Jeffrey WH . (1986). Elevated levels of microbial activity on the coral surface microlayer. Mar Ecol Prog Ser 33: 29–40.
Pfeffer W . (1884). Lokomotorische Richtungsbewgungen durchchemische Reize. Untersuchungen aus dem Botanischen Institut Tubingen 1: 363–482.
Raina JB, Tapiolas D, Willis BL, Bourne DG . (2009). Coral associated bacteria and their role in the biogeochemical cycling of sulfur. Appl Environ Microbiol 75: 3429–3501.
Raina JB, Dinsdale EA, Willis BL, Bourne DG . (2010). Do the organic sulphur compounds DMSP and DMS drive coral microbial associations? Trends Microbiol 18: 101–108.
Raina JB, Tapiolas DM, Foret S, Lutz A, Abrego D, Ceh J et al. (2013). DMSP biosynthesis by an animal and its role in coral thermal stress response. Nature 502: 677–682.
Rohwer F, Breitbart M, Jara J, Azam F, Knowlton N . (2001). Diversity of bacteria associated with the Caribbean coral Montastraea franksi. Coral Reefs 20: 85–91.
Rohwer F, Seguritan V, Azam F, Knowlton N . (2002). Diversity and distribution of coral-associated bacteria. Mar Ecol Prog Ser 243: 1–10.
Rohwer F, Kelly S . (2004). Culture independent analyses of coral-associated microbes. In Rosenberg E, Loya Y (eds) Coral Health and Disease. Springer: New York, NY, USA, pp 265–277.
Rosenberg E, Koren O, Reshef L, Efrony R, Zilber-Rosenberg I . (2007). The role of microorganisms in coral health, disease and evolution. Nat Rev 5: 355–362.
Salmond J, Loder J, Passenger J, Phinn S, Roelfsema C, Rempel C . (2013). Reef check Australia 2013 Heron Island Reef Healthy Report. Reef Check Foundation Ltd: Brisbane, Australia.
Schweinitzer T, Josenhans C . (2010). Bacterial energy taxis: a global strategy? Arch Microbiolo 192: 507–520.
Seymour JR, Seuront L, Mitchell JG . (2007). Microscale gradients of planktonic microbial communities above the sediment surface in a mangrove estuary. Estuar Coast Shelf Sci 73: 651–666.
Seymour JR, Ahmed T, Marcos, Stocker R . (2008). A microfluidic chemotaxis assay to study microbial behaviour in diffusing nutrient patches. Limnol Oceanogr Methods 6: 477–488.
Seymour JR, Marcos, Stocker R . (2009). Resource patch formation and exploitation throughout the marine microbial food web. Am Nat 173: e15–e29.
Seymour JR, Ahmed T, Simo R, Stocker R . (2010). Chemoattraction to DMSP in the marine microbial foodweb. Science 329: 342–34.
Sharp KH, Distal D, Paul VJ . (2012). Diversity and Dynamics of bacterial communities in early life stages of the Caribbean coral Porites astreoides. ISME J 6: 790–801.
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504.
Shnit-Orland M, Kushmaro A . (2009). Coral mucus-associated bacteria: a possible first line of defence. FEMS Microbiol Ecol 67: 371–380.
Siboni N, Ben-Dov E, Sivan A, Kushmaro A . (2008). Global distribution and diversity of coral-associated Archaea and their possible role in the coral holobion nitrogen cycle. Environ Microbiol 10: 2979–2990.
Smoot M, Ono K, Ruscheinski J, Wna PL, Ideker T . (2011). Cytoscape 2.8: new features for data integration and network visualisation. Bioinformatics 27: 431–432.
Stocker R, Seymour JR, Samadani A, Hunt DE, Polz MF . (2008). Rapid chemotactic response enables marine bacteria to exploit ephemeral microscale nutrient patches. Proc Natl Acad Sci USA 105: 4209–4214.
Stocker R, Seymour JR . (2012). Ecology and physics of bacterial chemotaxis in the ocean. Microbiol Mol Biol Rev 76: 792–812.
Sweet MJ, Croquer A, Bythell JC . (2011). Bacterial assemblages differ between compartments within the coral holobiont. Coral Reefs 30: 39–52.
Thomas WH, Owen RW Jr . (1971). Estimating phytoplankton production from ammonium and chlorophyll concentrations in nutrient-poor water of the eastern tropical pacific ocean. Fish Bull 69: 87–92.
Tout J, Jeffries TC, Webster NS, Stocker R, Ralph PJ, Seymour JR . (2014). Variability in microbial community composition and function between different niches within a coral reef. Microb Ecol 67: 540–552.
Vega Thurber R, Willner-Hall D, Rodriguez-Mueller B, Desnues C, Edwards RA, Angly F et al. (2009). Metagenomic analysis of stressed coral holobionts. Environ Microbiol 11: 2148–2163.
Vidal-Dupiol J, Ladriere O, Meistertzheim A-L, Foure L, Adjeroud M, Mitta G . (2011). Physiological responses of the scleractinina coral Pocillopora damicornis to bacterial stress from Vibrio coralliilyticus. J Exp Biol 214: 1533–1543.
Von Holt C, Von Holt M . (1968). The secretion of organic compounds by zooxanthellae isolated from various types of Zoanthus. Comp Biochem Physiol 24: 83–92.
Wadhams GH, Armitage JP . (2004). Making sense of it all: bacterial chemotaxis. Nat Rev 5: 1024–1037.
Wallace C . (1999) Staghorn Corals of the World: a Revision of the Genus Acropora. CSIRO: Collingwood, Australia.
Whitesides GM, Ostuni E, Takayama S, Jiang X, Ingber DE . (2001). Soft lithography in biology and biochemistry. Ann Rev Biomed Eng 3: 335–373.
Wild C, Huettel M, Klueter A, Kremb SG, Rasheed MYM, Jorgensen B . (2004). Coral mucus functions as an energy carrier and particle trap in the reef ecosystem. Nature 428: 66–70.
Wild C, Holger W, Huettel M . (2005). Influence of coral mucus on nutrient fluxes in carbonate sands. Mar Ecol Prog Ser 287: 87–98.
Wild C, Naumann M, Niggl W, Haas A . (2010). Carbohydrate composition of mucus released by sclearactinian warm- and cold-water reef corals. Aquat Biol 10: 41–45.
Acknowledgements
This research was funded by the Australian Research Council Grant DP110103091 to JRS, GWT and RS and the Human Frontiers in Science Program award no. RGY0089 to RS and JRS. This research was funded in part by the Gordon and Betty Moore Foundation through Grant #3801 to JRS, RS and GWT and an Investigator Award (grant GBMF3783) to RS; a post-graduate award to JT from the Department of Environmental Science and Climate Change Cluster at the University of Technology Sydney. JRS and NSW were funded through Australian Research Council Future Fellowships FT130100218 and FT120100480, respectively. We are grateful to the Great Barrier Reef Marine Park Authority for coral collection permits G09/31733.1 (PJR, University of Technology, Sydney). Amplicons were sequenced using the 454 GS-FLX pyrosequencing platform (Roche) at the Australian Centre for Ecogenomics (University of Queensland, Australia). Metagenomic sequencing was carried out at the Ramaciotti Centre for Gene Function Analysis at the University of New South Wales. The metagenomes can be accessed through MG-Rast (http://metagenomics.anl.gov) under the project numbers 4483104.3 (A. palifera in the lagoon), 4483105.3 (A. aspera on the reef crest), 4483106.3 (sandy substrate) and 4483107.3 (open water).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Tout, J., Jeffries, T., Petrou, K. et al. Chemotaxis by natural populations of coral reef bacteria. ISME J 9, 1764–1777 (2015). https://doi.org/10.1038/ismej.2014.261
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2014.261
This article is cited by
-
Genome analysis of a coral-associated bacterial consortium highlights complementary hydrocarbon degradation ability and other beneficial mechanisms for the host
Scientific Reports (2023)
-
Novel marine metalloprotease—new approaches for inhibition of biofilm formation of Stenotrophomonas maltophilia
Applied Microbiology and Biotechnology (2023)
-
Benthic exometabolites and their ecological significance on threatened Caribbean coral reefs
ISME Communications (2022)
-
Coral holobiont cues prime Endozoicomonas for a symbiotic lifestyle
The ISME Journal (2022)
-
Vibrio tetraodonis subsp. pristinus subsp. nov., isolated from the coral Acropora cytherea at Palmyra Atoll, and creation and emended description of Vibrio tetraodonis subsp. tetraodonis subsp. nov
Antonie van Leeuwenhoek (2022)