Abstract
Chemotaxis allows microorganisms to rapidly respond to different environmental stimuli; however, understanding of this process is limited by conventional assays, which typically focus on the response of single axenic cultures to given compounds. In this study, we used a modified capillary assay coupled with flow cytometry and 16S rRNA gene amplicon pyrosequencing to enumerate and identify populations within a lake water microbial community that exhibited chemotaxis towards ammonium, nitrate and phosphate. All compounds elicited chemotactic responses from populations within the lake water, with members of Sphingobacteriales exhibiting the strongest responses to nitrate and phosphate, and representatives of the Variovorax, Actinobacteria ACK-M1 and Methylophilaceae exhibiting the strongest responses to ammonium. Our results suggest that chemotaxis towards inorganic substrates may influence the rates of biogeochemical processes.
Similar content being viewed by others
Main
In aquatic environments, organic and inorganic nutrients are distributed heterogeneously, with microscale hotspots found throughout the water column (Azam, 1998; Blackburn et al., 1998). Microbial cells are generally unable to uptake nutrients that are more than a few cell diameters away; therefore, motility is a key determinant of the size of the microhabitats within which different populations perceive and exploit nutrient sources (Stocker, 2012). Non-motile cells can explore ∼80 nl water per day, which when compared with ca. 1 ml per day for motile cells is very small (Stocker, 2012). Furthermore, as motility is typically associated with the ability of cells to sense and direct movement along chemical gradients (chemotaxis), many motile populations are not only able to explore large volumes of water, but are also able to move directly towards perceived resource hotspots (Fenchel, 2002; Stocker et al., 2008; Stocker and Seymour, 2012).
Aquatic bacterial chemotaxis occurs in response to a wide range of organic substrates, including amino acids (Barbara and Mitchell, 2003), sugars (Malmcrona-Friberg et al., 1990) and organic sulphur compounds (Miller et al., 2004). In contrast, chemotaxis towards inorganic substrates is poorly characterised despite bacterial growth in aquatic ecosystems often being limited by dissolved inorganic nutrients (Church, 2008), and the requirement for some inorganic chemicals in anaerobic respiration (Thauer et al., 1977). In addition, as chemotaxis studies have focussed on single populations of cultivated bacteria, the generality of this trait is unknown. To date, chemotaxis towards inorganic substrates has been observed towards: (1) ammonium and nitrate by the marine cyanobacterium, Synechococcus (Willey and Waterbury, 1989); (2) ammonium by the purple non-sulphur photosynthetic bacterium Rhodobacter sphaeroides (Ingham and Armitage, 1987; Poole and Armitage, 1989); (3) nitrate and nitrite by Shewanella putrefaciens (Nealson et al., 1995) and several strains of denitrifying bacteria (Kennedy and Lawless, 1985; Lee et al., 2002) and (4) phosphate by a heterotrophic marine Thalassospira sp. (Hütz et al., 2011).
In this study, we used a syringe-based assay (Supplementary Figure S1), inspired by the traditional capillary assay (Adler and Dahl, 1967), coupled with flow cytometry and 16S rRNA gene amplicon pyrosequencing to enumerate and identify populations within a lake water microbial community that exhibited chemotaxis towards ammonium, nitrate and phosphate. Chemotaxis assays were performed by submerging 1-ml syringes containing 80 μl of 0.1 M chemoattractant (nitrate/ammonium/phosphate) into lake water for 30 min (Supplementary information). Each chemoattractant was prepared in 0.2 μm-filtered lake water to ensure that the chemical characteristics of the background solution in the syringe was similar to that in the lake. Filtered lake water control assays with no added chemoattractant were performed in parallel and facilitated the measurement of stochastic movement of cells into our assays (Supplementary Figure S2). All assays were replicated six times, providing three samples for cell counting and three samples that were pooled for 16S rRNA gene amplicon sequencing. This novel approach facilitated culture-independent characterisation of chemotactic populations within a lake water community. To ensure that counts in the assays reflected chemotaxis rather than growth stimulation, we enumerated cells in 0.2 μm-filtered lake water with and without added nutrients. The controls were incubated for 30 min, but the tips of the syringes were not submerged in lake water (Supplementary Figure S3).
Cell counts in the nutrient-spiked chemotaxis assays were approximately six times higher than in the control assays (P<0.001, generalized linear model; Supplementary Figures S2 and S3), but did not differ from one another. Cell counts in nutrient stimulation assays, however, did not differ from those associated with the filtered lake water control (Supplementary Figure S3). These results indicate that nitrate, ammonium and phosphate elicited strong chemotactic responses from at least one population within the lake water community and that slight differences in the diffusion coefficients of the attractants (ca. 0.17, 0.18 and 0.05 × 104 cm2 s−1 for NO3−, NH4+ and PO43−, respectively) did not lead to differences in the strength of response.
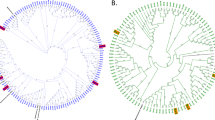
Three members of the Sphingobacteriales exhibited the strongest chemotactic response to nitrate and phosphate (Figures 1 and 2). Phylogenetic analysis (Supplementary information) revealed that these Sphingobacteriales populations belonged to the family Chitophagaceae. Within this family, members of the genera Niastella and Chitinophaga are known to exhibit gliding motility (Lim et al., 2009; Weon et al., 2009; Del Rio et al., 2010), and some Chitinophaga species are known to reduce nitrate as a terminal electron acceptor (Lim et al., 2009). Interestingly, the only other Sphingobacteriales population observed in the lake water at >1% relative abundance, a Pedobacter-like organism, did not exhibit chemotaxis towards nitrate (Figure 1). This is consistent with observations that Pedobacter isolates do not reduce nitrate (Steyn et al., 1998). Therefore, the positive chemotactic response towards nitrate observed for the Sphingobacteriales populations may reflect their ability to detect and exploit nitrate as an alternative electron acceptor. Chemoattraction may also reflect the ability for cells to move towards nitrate in response to inorganic nutrient deficiency; however, there are examples, where bacteria do not metabolise attractants. For example, Escherichia coli is attracted to, but does not metabolise methyl-aspartate (Adler, 1969).
Bacterial operational taxonomic units (OTUs) present at >1% abundance in any sample. The size of each circle represents the relative abundance of each OTU normalised by the total cell count within each sample. The OTUs are organised by phylogenetic relatedness based on a maximum likelihood bootstrapped tree (1000 iterations) of full-length sequences that were identified as the nearest BLAST matches for each OTU. Bootstrap values are represented as follows: 75–100% (closed circles), 50–74% (open circles), <50% (no circle). The compositional similarity of each community is represented at the base of the heatmap by a complete-linkage cluster analysis tree of OTU abundances.
PCA ordination highlighting differences in operational taxonomic units (OTU) abundance between the chemotaxis assays. OTU abundances were calculated by normalising the relative abundances of each OTU by the total cell count within each sample. The OTUs are represented as crosses, with the taxonomic affiliation of the most discriminating OTUs highlighted in text.
Our results indicate that chemotaxis towards inorganic phosphate is exhibited by a range of bacteria, in particular, the Sphingobacteriales, which also showed high levels of chemotaxis towards nitrate (Figures 1 and 2). Previously, chemoattraction to phosphate has only been demonstrated in a free-living planktonic bacterium belonging to the genus Thalassopira (Hütz et al., 2011), which was isolated from the oligotrophic eastern Mediterranean Sea. Interestingly, no differences in the swimming speed of the Thalassopira species was observed when exposed to organic carbon or inorganic phosphate (Hütz et al., 2011), suggesting that its motility was not differentially regulated by the type of nutrient supplied. This observation is consistent with our data, which showed that the populations responding to nitrate were similar to those that respond to phosphate (Figures 1 and 2). This may be indicative of cross-regulation of chemotaxis machinery by nitrate and phosphate sensory systems or environmental co-dependency on these compounds. Our results substantially expand the known diversity of bacteria that exhibit chemotaxis towards phosphate.
Ammonium elicited the strongest response among the tested inorganic nutrients for the majority of dominant populations, although none of these responses were as strong as those observed for nitrate and phosphate by the Sphingobacteriales (Figure 1). Organisms closely related to Variovorax, Actinobacteria ACK-M1, also known as the acI group (Warnecke et al., 2004), and a member of Methylophilaceae exhibited the strongest responses to ammonium (Figures 1 and 2). Variovorax paradoxus has been previously reported to swarm in response to ammonium (Jamieson et al., 2009), and ammonium is known to influence the composition of Actinobacteria ACK-M1 assemblages in lakes (Newton et al., 2007); however, we are not aware of studies demonstrating that representatives of the Methylophilaceae and Actinobacteria ACK-M1 group exhibit chemotaxis towards ammonium. This lack of information highlights that our approach has the potential to greatly improve knowledge of the microbial lineages that exhibit chemotaxis towards a wide range of compounds in natural environments. Our assay should also facilitate functional characterisation of assemblages of chemotactic populations when coupled with metagenomics and metatranscriptomics. This approach would help to reveal the range and prevalence of mechanisms of movement and environmental sensing used by chemotactic populations in natural environments. Gliding motility, for example, is used by Myxococcus xanthus during chemotaxis to phosphatidylethanolamine (Kearns and Shimkets, 1998), but it is unknown whether this type of motility is common among chemotactic microorganisms. Similarly, much of what is known about how microbial chemoreceptors respond to changes in concentrations of chemical attractants or repellents is based on E. coli and Salmonella typhimurium (Falke and Hazelbauer, 2001). Current evidence indicates, however, that chemosensory systems that mediate microbial chemotaxis differ considerably between species (Montrone et al., 1998; Boin et al., 2004; Szurmant and Ordal, 2004; Meier et al., 2007). The extent of these knowledge gaps highlights that our understanding of chemotaxis in natural environments is far from complete.
Our study indicates that a wide range of lake water bacterial populations have the capacity to move towards inorganic nutrients. This observation was previously overlooked due to culture-based analyses. Chemotaxis towards inorganic nutrients is a behaviour that likely enables them to exploit microscale gradients of limiting resources in the environment. It has previously been suggested that bacterial chemotactic exploitation of microscale patches of organic compounds in marine habitats can influence carbon cycling processes and influence microbial community interactions (Azam, 1998; Blackburn et al., 1998; Fenchel, 2002). Our observations indicate that strong, but phylogenetically variable, chemotactic responses towards inorganic compounds may similarly influence nutrient cycling processes and microbial competitive interactions within aquatic ecosystems.
References
Adler J . (1969). Chemoreceptors in bacteria. Science 166: 1588–1597.
Adler J, Dahl MM . (1967). A method for measuring the motility of bacteria and for comparing random and non-random motility. J Gen Microbiol 46: 161–173.
Azam F . (1998). Microbial control of oceanic carbon flux: the plot thickens. Science 280: 694–696.
Barbara GM, Mitchell JG . (2003). Marine bacterial organisation around point-like sources of amino acids. FEMS Microbiol Ecol 43: 99–109.
Blackburn N, Fenchel T, Mitchell JG . (1998). Microscale nutrient patches in plankton habitats shown by chemotactic bacteria. Science 282: 2254–2256.
Boin MA, Austin MJ, Hase CC . (2004). Chemotaxis in Vibrio cholerae. FEMS Microbiol Lett 239: 1–8.
Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R . (2010). PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26: 266–267.
Church MJ . (2008). Resource control of bacterial dynamics in the sea. In: Kirchman D (ed). Microbial Ecology of the Ocean. 2nd edition, John Wiley & Sons, Inc: Hoboken, NJ, USA.
Del Rio TG, Abt B, Spring S, Lapidus A, Nolan M, Tice H et al (2010). Complete genome sequence of Chitinophaga pinensis type strain (UQM 2034T). Stand Genomic Sci 2: 87–95.
Falke JJ, Hazelbauer GL . (2001). Transmembrane signaling in bacterial chemoreceptors. Trends Biochem Sci 26: 257–265.
Fenchel T . (2002). Microbial behaviour in a heterogenous world. Science 296: 1068–1071.
Hütz A, Schubert K, Overmann J . (2011). Thalassospira sp. isolated from the oligotrophic eastern Mediterranean Sea exhibits chemotaxis toward inorganic phosphate during starvation. Appl Environ Microbiol 77: 4412–4421.
Ingham CJ, Armitage JP . (1987). Involvement of transport in Rhodobacter sphaeroides. J Bacteriol 169: 5801–5807.
Jamieson WD, Pehl MJ, Gregory GA, Orwin PM . (2009). Coordinated surface activities in Variovorax paradoxus EPS. BMC Microbiol 9: 124.
Kearns DB, Shimkets LJ . (1998). Chemotaxis in a gliding bacterium. Proc Natl Acad Sci USA 95: 11957–11962.
Kennedy MJ, Lawless JG . (1985). Role of chemotaxis in the ecology of denitrifiers. Appl Environ Microbiol 49: 109–114.
Lee DY, Ramos A, Macomber L, Shapleigh JP . (2002). Taxis response of various denitrifying bacteria to nitrate and nitrite. Appl Environ Microbiol 68: 2140–2147.
Lim JH, Baek SH, Lee ST . (2009). Ferruginibacter alkalilentus gen. nov., sp. nov. and Ferruginibacter lapsinanis sp. nov., novel members of the family ‘Chitinophagaceae’ in the phylum Bacteroidetes, isolated from freshwater sediment. Int J Syst Evol Microbiol 59: 2394–2399.
Malmcrona-Friberg K, Goodman A, Kjelleberg S . (1990). Chemotactic responses of marine Vibrio sp. Strain S14 (CCUG 15956) to low-molecular weight substances under starvation and recovery conditions. Appl Environ Microbiol 56: 3699–3704.
Meier VM, Muschler P, Scharf BE . (2007). Functional analysis of nine putative chemoreceptor proteins in Sinorhizobium meliloti. J Bacteriol 189: 1816–1826.
Miller TR, Hnilicka K, Dziedzic A, Desplats P, Belas R . (2004). Chemotaxis of Silicibacter sp. strain TM1040 toward dinoflagellate products. Appl Environ Microbiol 70: 4662–4701.
Montrone M, Eisenbach M, Oesterhelt D, Marwan W . (1998). Regulation of switching frequency and bias of the bacterial flagellar motor by CheY and fumarate. J Bacteriol 180: 3375–3380.
Nealson KH, Moser DP, Saffarini DA . (1995). Anaerobic electron acceptor chemotaxis in Shewanella putrefaciens. Appl Environ Microbiol 61: 1551–1554.
Newton RJ, Jones SE, Helmus MR, McMahon KD . (2007). Phylogenetic ecology of the freshwater Actinobacteria acI lineage. Appl Environ Microbiol 73: 7169–7176.
Poole PS, Armitage JP . (1989). Role of metabolism in the chemotactic response of Rhodobacter sphaeroides to ammonia. J Bacteriol 171: 2900–2902.
Steyn PL, Segers P, Vancanneyt M, Sand P, Kersters K, Joubert JJ . (1998). Classification of heparinolytic bacteria into a new genus, Pedobacter, comprising four species: Pedobacter heparinus comb. nov., Pedobacter piscium comb. nov., Pedobacter africanus sp. nov. and Pedobacter saltans sp. nov. Proposal of the family Sphingobacteriaceae fam. nov. Int J Syst Bacteriol 48: 165–177.
Stocker R . (2012). Marine microbes see a sea of gradients. Science 338: 628–633.
Stocker R, Seymour JR . (2012). Ecology and physics of bacterial chemotaxis in the ocean. Microbiol Mol Biol Rev 76: 792–812.
Stocker R, Seymour JR, Samadan A, Hunt DH, Polz M . (2008). Rapid chemotactic response enables marine bacteria to exploit ephemeral microscale nutrient patches. Proc Natl Acad Sci USA 105: 4209–4214.
Szurmant H, Ordal GW . (2004). Diversity in chemotaxis mechanisms among the bacteria and archaea. Microbiol Mol Biol Rev 68: 301–319.
Thauer RK, Jungerman K, Decker K . (1977). Energy conservation in chemotrophic anaerobic bacteria. Bacteriol Rev 41: 100–180.
Warnecke F, Amann R, Pernthaler J . (2004). Actinobacterial 16S rRNA genes from freshwater habitats cluster in four distinct lineages. Environ Microbiol 6: 242–253.
Weon HY, Yoo SH, Kim BY, Son JA, Kim YJ, Kwon SW . (2009). Niabella ginsengisoli sp. nov., isolated from soil cultivated with Korean ginseng. Int J Syst Evol Microbiol 59: 1282–1285.
Willey JM, Waterbury JB . (1989). Chemotaxis toward nitrogenous compounds by swimming strains of marine Synechococcus spp. Appl Environ Microbiol 55: 1888–1894.
Acknowledgements
We thank Philip Hugenholtz for advice on the manuscript. PGD acknowledges financial support from the Commonwealth Scientific and Industrial Research Organisation. GWT is supported by an ARC Queen Elizabeth II fellowship (DP1093175).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Dennis, P., Seymour, J., Kumbun, K. et al. Diverse populations of lake water bacteria exhibit chemotaxis towards inorganic nutrients. ISME J 7, 1661–1664 (2013). https://doi.org/10.1038/ismej.2013.47
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2013.47
Keywords
This article is cited by
-
Characterization of two keystone taxa, sulfur-oxidizing, and nitrate-reducing bacteria, by tracking their role transitions in the benzo[a]pyrene degradative microbiome
Microbiome (2023)
-
Indole decreases the virulence of the bivalve model pathogens Vibrio tasmaniensis LGP32 and Vibrio crassostreae J2-9
Scientific Reports (2022)
-
Dispersal-competition tradeoff in microbiomes in the quest for land colonization
Scientific Reports (2018)
-
Selective isolation and characterisation of novel members of the family Nocardiopsaceae and other actinobacteria from a marine sediment of Tioman Island
Antonie van Leeuwenhoek (2018)
-
Brown Rot Syndrome and Changes in the Bacterial Сommunity of the Baikal Sponge Lubomirskia baicalensis
Microbial Ecology (2018)