Abstract
The association of phytoplankton with bacteria is ubiquitous in nature and the bacteria that associate with different phytoplankton species are very diverse. The influence of these bacteria in the physiology and ecology of the host and the evolutionary forces that shape the relationship are still not understood. In this study, we used the Pseudo-nitzschia–microbiota association to determine (1) if algal species with distinct domoic acid (DA) production are selection factors that structures the bacterial community, (2) if host-specificity and co-adaptation govern the association, (3) the functional roles of isolated member of microbiota on diatom–hosts fitness and (4) the influence of microbiota in changing the phenotype of the diatom hosts with regards to toxin production. Analysis of the pyrosequencing-derived 16S rDNA data suggests that the three tested species of Pseudo-nitzschia, which vary in toxin production, have phylogenetically distinct bacterial communities, and toxic Pseudo-nitzschia have lower microbial diversity than non-toxic Pseudo-nitzschia. Transplant experiments showed that isolated members of the microbiota are mutualistic to their native hosts but some are commensal or parasitic to foreign hosts, hinting at co-evolution between partners. Moreover, Pseudo-nitzschia host can gain protection from algalytic bacteria by maintaining association with its microbiota. Pseudo-nitzschia also exhibit different phenotypic expression with regards to DA production, and this depends on the bacterial species with which the host associates. Hence, the influences of the microbiota on diatom host physiology should be considered when studying the biology and ecology of marine diatoms.
Similar content being viewed by others
Introduction
Every eukaryotic organism has its own microbiota, either found as epibionts or endobionts in a living host, which exert tremendous physiological and developmental influences on the host. Even small phytoplankton such as diatoms are no exception to this paradigm. Diatoms are the most prolific primary producers in the ocean and contribute to one-fifth of the primary productivity on earth (Falkowski et al., 1998). Diatoms form algal blooms in the coastal and open ocean, where they sequester carbon in deep sediments, a major influence in the global carbon cycle (Armbrust, 2009). Some bloom-forming diatoms are toxic, such as members of the cosmopolitan genus Pseudo-nitzschia (Bates et al., 1989). Of the 37 described species, 12 produce the neurotoxin domoic acid (DA) (Trainer et al., 2012). This compound has been found in zooplankton, shellfish, crustaceans, echinoderms, worms, marine mammals, marine birds and in sediments, with subsequent transfer to benthic organisms such as commercially harvested flatfish and crabs (Trainer et al., 2012). DA, which poses a threat to ocean and human health, also brings monetary losses from temporary closure of shellfish aquaculture, seafood and tourism industries worldwide (Hoagland et al., 2002; Trainer et al., 2007). Human intoxication is referred to as amnesic shellfish poisoning; clinical signs in humans include gastrointestinal distress, confusion, disorientation, seizures, permanent short-term memory loss and in severe cases, death (Perl et al., 1990).
-
First discovered as a human health problem in 1987 after causing illness and death in Prince Edward Island, Canada (Bates et al., 1989), there is still no clear consensus on the ecological significance or triggers for DA production. Toxicity has been related to nutrient stress, light, salinity, trace metals (particularly iron and copper) and more generally, as a stress response or in association with slowing or cessation of growth (see review by Trainer et al., 2012). The toxin is most often found intracellularly, but significant quantities can also be released to the media (Wells et al., 2005), leading to hypotheses about the role of DA in modulating the associated heterotrophic bacterial assemblage (Bates et al., 1995; Guannel et al., 2011; Trainer et al., 2012).
-
Although limited in study, the interaction of diatoms with bacteria is suggested to have an important role in their ecological success, such as the promotion of algal blooms (see review by Doucette, 1995). More than 5% of the diatom genome consists of genes derived from bacteria (Armbrust et al., 2004; Bowler et al., 2008), attesting to the evolutionary history of interaction between the two. Bacteria and phytoplankton interact at the ‘phycosphere’, where the bacteria can access the algal exudates (Bell and Mitchell 1972). Among diatoms, the bacterial associates are found underneath the cingulae, raphe and above the poroids of striae (Kaczmarska et al., 2005). Host exudates like polysaccharides, small amino acids, sugars, proteoglycans or glycoproteins, can be species-specific (Myklestad, 1995) and may govern the kinds of bacteria that associate with a diatom species (Sapp et al., 2007). Moreover, the species, growth and the physiological state of the algae may govern the succession of the attached bacterial community (Grossart et al., 2005). Specific effects of associated bacteria on diatom physiology include increasing diatom aggregation and sinking rate as in the case of Thalassiosira (Gärdes et al., 2010), and toxin production such as that seen in the diatom Pseudo-nitzschia (Bates et al., 1995). Associated bacteria also have differential influence in the secretion of polysaccharides when co-cultured with a biofilm-forming freshwater diatom, Cymbella microcephalla (Bruckner et al., 2008). The cyanobacterial production of the high toxic form of microcystin is also increased when Microcystis aeruginosa is co-cultured with heterotrophic bacteria at increasing temperature (Dziallas and Grossart, 2011a). Interestingly, associated bacteria can also degrade microcystin under laboratory conditions (Dziallas and Grossart, 2011b).
The dominant paradigm for bacteria–phytoplankton interaction is that phytoplankton serves as organic nutrient sources for saprophytic bacteria as depicted in the classical idea of microbial loop (Azam et al., 1983; Azam 1998). This paradigm is strengthened by reports of marine bacteria that prey on living phytoplankton (reviewed in Doucette et al., 1998 and Mayali and Azam 2004). On the contrary, bacteria can also be mutualistic to phytoplankton (Cole 1982; Mouget et al., 1995; Stewart et al., 1997; Grossart 1999; Ferrier et al., 2002; Liu et al., 2008; Amin et al., 2009). Although these opposing interactions of diatom and bacteria are both observed in nature, there is currently a lack of cohesive evolutionary principle that explains these interactions.
In this study, we investigated these algae–bacteria relationships in the light of microbiota framework under defined laboratory conditions. We considered all attached bacteria as symbionts regardless of their function (mutualistic, parasitic or commensal) (Ebert and Qi, 2010). We asked if evolutionary forces such as host-specificity and co-adaptation govern the interactions between marine diatoms and their microbiota. We chose the genus Pseudo-nitzschia for this study because DA in Pseudo-nitzschia species, along with fitness measurement, allow us to evaluate the influence of microbiota on diatom host physiology. The genome and transcriptomes of several Pseudo-nitzschia species are also available (genome: http://genome.jgi.doe.gov/Psemu1/Psemu1.download.html; transcriptomes: http://genomeportal.jgi.doe.gov/PsenittraphaseII/PsenittraphaseII.download.html and https://www.marinemicroeukaryotes.org/project_organisms?direction=desc&page=9&per_page=25&sort=organisms.genus_name), which provide a reference for identifying the influences of bacteria on Pseudo-nitzschia physiology at the molecular level and placing these findings in an ecologically relevant framework. At present, not much is known about the interaction of bacteria with Pseudo-nitzschia. Using culture method, Kaczmarska et al. (2005) reported that the bacteria associating with the toxic Pseudo-nitzschia are very diverse and bacterial morphotype succession varies depending on the algal growth stage. ARISA screening of toxigenic and non-toxigenic Pseudo-nitzschia suggest that Pseudo-nitzschia species harbor distinct bacterial associates (Guannel et al., 2011). A foreign bacterium is reported to elevate DA production in Pseudo-nitzschia multiseries (Bates et al., 1995), while some bacteria may utilize DA (Stewart, 2008). In this study, we took a three-step approach to address our primary questions. First, we evaluated whether the bacterial communities of Pseudo-nitzschia species with different DA production are distinct from each other using pyrosequencing approach. This is to better characterize the diversity of the bacterial community, which would include the unculturable members of the microbiome. Next, we determined the functional role of isolated members of the microbiota on the diatom hosts’ fitness by transplant experiments. Lastly, we examined which bacterial member of the microbiota could alter the DA production of a clonal Pseudo-nitzschia.
Materials and methods
Algal and bacterial isolation, culture and molecular genotyping
Single Pseudo-nitzschia cell isolates were obtained from bloom seawater samples collected from the Santa Cruz Wharf, California. Single cells were picked with a sterile micropipette and washed 10 times with filter-sterilized seawater. Cells were grown first in F/10-Se media. Once cells proliferated, the cultures were maintained in F/2-Se (Guillard and Ryther, 1962) medium under growth conditions of 12:12 photoperiod, 15 °C and 80 μE m−2 in an algal incubator. The Pseudo-nitzschia cultures were identified by morphology using light microscopy and genotyped by sequencing the 18S rRNA gene (see Supplementary Information for genotyping methods).
The microbiota of Pseudo-nitzschia pungens (PP) and Pseudo-nitzschia australis (PA) were obtained by filtering healthy algal cells onto sterile 5 μm filters (Whatman, Dassel, Germany) and washed once with filter-sterilized seawater. The filters were dabbed on the surface of marine agar (Difco 2216, BD, Franklin Lakes, NJ, USA) several times and incubated in the algal incubator until bacterial colonies were observed. Pure single colonies were obtained by repeated streaking in agar medium and grown in 5-ml marine broth (Difco 2216) at room temperature for 48 h. Bacterial isolates were identified using 16S rDNA sequencing (see Supplementary Information). Stock cultures of the bacterial isolates were stored in 50/50 glycerol in −80 °C.
Sequences were analyzed using Seqman II (version 9.0 DNASTAR Inc, Madison, WI, USA) with minimum similarity set at 97%. Bacterial taxon for each sequence was identified and named by the homologous 16S sequence in Genbank using BLAST (Altschul et al., 1990). Names were appended with PP or PA to indicate source of algal host. Sequences accession numbers are listed in Table 1.
454 Pyrosequencing and statistical analysis
Pseudo-nitzschia cultures (P. australis=3, Pseudo-nitzschia fraudulenta=2, P. pungens=2) in 100 ml volume were harvested at late-exponential stage by centrifugation for 10 min at 3000 g. Pellets were stored in sterile TE buffer at −80 °C and sent to Research and Testing Laboratory (Lubbock, TX, USA) for pyrosequencing. Bacterial DNA extraction and 16S rDNA pyrosequencing were carried out using the protocol described in Dowd et al. (2008) and Sun et al. (2011) (see Supplementary Information). The bTEFAP method was based on the Titanium reagents and protocol for Genome Sequencer FLX System (Roche Indianapolis, IN, USA). The V6–V9 region of the 16S rDNA gene was targeted for the 454 pyrosequencing run using the universal primers Yellow939F and Yellow1492R.
Following sequencing, a quality check was performed to remove short sequence reads (length <150 bp), low quality sequences (score <25, Huse et al., 2007) and any non-bacterial ribosome sequences and chimeras (Gontcharova et al., 2010). Post-processing of the pyrosequencing output was carried out using the Quantitative Insights Into Microbial Ecology (QIIME 1.3) pipeline (Caporaso et al., 2010). Operational taxonomic units (OTUs) were picked based on clustering sequences at 97% similarity using Uclust (Edgar, 2010). Representative sequences picked from clustered OTUs were aligned using Pynast (Caporazo, Bittinger et al., 2010) with the Greengenes core alignment as a reference (DeSantis et al., 2006). Identification of OTUs was based on RDP classifier 2.2 (Wang et al., 2007), while the assignment of bacterial taxon was performed using BLAST (Altschul et al., 1990, E-value 10−20 (megablast only); minimum coverage 97%; minimum pairwise identity 90–97%.). Singletons and chloroplast sequences from the OTU table were filtered before bacterial diversity analyses. A phylogenetic tree of the OTUs was made with Fast Tree 2.1.3 (Price et al., 2010). The raw sequencing reads have been deposited at the NCBI Short Read Archive under BioProject ID PRJNA182211 with the following BioSample accession numbers: P. pungens-B2A (SAMN01818668), P. pungens-C5A (SAMN01818669), P. fraudulenta-1 (SAMN01818670), P. fraudulenta-8 (SAMN01818671), P. australis-12 (SAMN01818672), P. australis-15 (SAMN01818673), P. australis-B5 (SAMN01818674).
Microbial community comparison
The microbial diversity within each Pseudo-nitzschia sample was assessed using three alpha diversity indices (number of species, phylogenetic diversity and Shannon index). To control for sequencing effort, samples were rarified at a depth of 6650 sequences with 10 sequences at each step and at a depth of 151 sequences with 1 sequence step, performing 10 and 100 iterations at each sampling depth, respectively.
The similarity of bacterial communities between Pseudo-nitzschia species was compared by jackknifing the OTU tables, the distance matrices and the UPGMA tree clusters at 1000 iterations. To statistically test the phylogenetic differences of the microbial communities between samples, beta significance was determined using Unweighted UniFrac with Monte Carlo Significance Test at 1000 iterations.
Algal fitness assay
The Pseudo-nitzschia cultures were made axenic with a treatment of Ampicillin (1 mg ml−1) and Kanamycin (10 μg ml−1) for three culture generations. Axenicity was verified via sterility test, inoculating sterile marine broth with 0.5 ml of previously antibiotic-treated algal cultures for 72 h and by PCR screening of 16S rDNA using bacterial universal primers 27 F and 907R (5′-CCGTCAATTCMTTTRAGTT-3′). All tests were negative for bacterial presence.
A binary culture (axenic algae and single bacterium) experiment was set-up to assess the individual influence of the microbiota on algal fitness; one set was to test the effect of microbiota on native Pseudo-nitzschia hosts by re-introducing the bacterial isolates into the same host species; the other set was to test the effect of microbiota on a foreign host by transplanting the microbiota of one Pseudo-nitzschia species to another. All assays were carried on at least two hosts species strains except for P. fraudulenta (see Table 1). In a fresh 4-ml sterile F/2 medium in borosilicate tubes, 0.1 ml of healthy algae were co-cultured with 15 μl freshly harvested bacteria previously re-suspended in filter-sterilized seawater with an OD600 of 0.65–0.81 at a final bacterial density of 1.4 × 106–3.5 × 106 colony forming units (CFU) ml−1 in the co-cultures. Preparation of bacterial inoculum for this assay and DA assay is described in the Supplementary Section. Bacterial counts (in CFU) were obtained using the drop plate method (Hoben and Somasegaran, 1982). To determine the influence of bacteria on host fitness, algal fluorescence, which is a proxy for algal biomass, was measured daily with a fluorometer (Turner Quantech, Barnstead, Thermo Scientific, Waltham, MA, USA) and the biomass was used to estimate logarithmic growth. Algal specific growth (SPG) rates per day were calculated during exponential growth phase (between day 2–5 for all cultures, for consistency) using the equation: SPG=Ln (FIU2−FIU1)/t2−t1.
We also tested the fitness of non-axenic Pseudo-nitzschia against a bacterium that was algicidal to axenic P. australis and P. fraudulenta but mutualistic to P. pungens. We extended the fitness assay to three non-axenic strains of P. australis and P. fraudulenta following the methods described above. SPG rates per day were calculated between day 1–4.
DA measurement in binary culture
To test the hypothesis that bacteria can influence the toxin production of the Pseudo-nitzschia, the above fitness assay experiment was carried out on the same day in duplicate to triplicate 50-ml cultures and grown up to late-exponential phase (12–15 days). In a sterile tissue culture flask (Corning Inc., Tewksbury, MA, USA), 0.3 ml axenic healthy algae were added to the sterile F/2 medium and co-cultured with 50 μl of the same bacterial inocula used on the fitness assay above at a final bacterial density of 4.8 × 105–9.2 × 105 CFU ml−1 in the co-cultures. An aliquot was taken for algal density count in a Sedgewick-Rafter cell counter before algal harvest. Cultures were then filtered with 25 mm Whatman GF/C filters, and stored in −80 °C until processing. DA was extracted by adding 1.5 ml 20% methanol, sonicated on ice for 3 min, centrifuged for 10 mins at 3000 g and the supernatant filtered with a 0.2 μm syringe filter (Litaker et al., 2008). DA concentration was measured on a microplate reader (Spectromax M2, Molecular Devices, Sunnyvale, CA, USA) following the procedure described in Mercury Science DA kit (Mercury Science, Durham, NC, USA).
Statistical analysis
One-way analysis of variance was used to analyze the differences in algal growth rates of Pseudo-nitzschia co-cultured with different bacteria. Data were checked for normality and heterogeneity of variances before analysis. Post-hoc means were compared with Student’s t-test and only P-values against axenic culture were reported from the post-hoc mean comparisons. To determine the influence of bacteria on DA content of native and foreign Pseudo-nitzschia hosts, analysis of the means transformed ranks with the α level set at 0.05 was used to compare sample means from the group means. All statistical analyses were done in JMP Pro 10.0. (SAS Institute, Cary, NC, USA). Means and standard errors were reported.
Results
Pseudo-nitzschia species harbor distinct and diverse microbial communities
Genotyping by 18S rRNA sequencing confirmed the identity of three species of Pseudo-nitzschia in this study. They are P. pungens, P. fraudulenta and P. australis (see Table 1). DA tests in early stationary phase showed that P. fraudulenta and P. australis produced DA at 14.8 and 22.7 μg l−1 and 137.7 and 179.4 μg l−1 in non-axenic and axenic state, respectively, while P. pungens did not produce DA and is considered non-toxic (see also Figure 5 and Supplementary Figure S5).
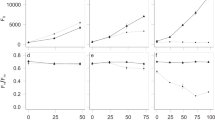
Cellular DA concentration (femtogram per cell) measured from P. australis (a) and P. fraudulenta (b) at late-exponential stage after co-cultivation with individual bacteria isolated from P. pungens and P. australis. PA, P. australis; PP, P. pungens; AX, axenic (n=3; 3); NON-AX, non-axenic (n=3; 2); GLA, Glaciecola (n=2; 3); MAR, Marinobacter (n=3; 2); PSE, Pseudoalteromonas (n=2; 3); ALT, Alteromonas (n=3; 3); ROSEO, Roseobacter (n=3; 2); PHAEO, Phaeobacter (n=2; 2); SAL, Salegentibacter (n=3; 2); CEL, Cellulophaga (n=3; 2); POL, Polaribacter (n=2; 2); PLAN, Planococcus (n=3; 2); BACI, Bacillus (n=3; 3). The group mean (line), sample mean (midline inside diamond), mean error bars and the 95% confidence points for each group represented by the diamond’s top and bottom points are depicted in the graph. Asterisks denote significant differences (P<0.05) between bacterial treatment and group mean by analysis of the Means with transformed ranks. Number of trials, n, for P. australis and P. fraudulenta, respectively, are indicated in parenthesis after name of bacteria with each bacterial treatment.
Pyrosequencing of microbiota associating with the three Pseudo-nitzschia species yielded a total of 91 701 qualified reads (Table 2) that were clustered to a total of 439 OTUs. Singletons and diatom chloroplast sequences were removed leaving 266 OTUs for the microbial diversity and clustering analyses.
At an even depth of 151 sequences, the non-toxic P. pungens samples showed higher microbial diversity than P. fraudulenta and P. australis based on phylogenetic diversity, observed species and Shannon diversity indices. At a sequence depth of 6650, the toxic species had higher diversity than P. pungens when using the phylogenetic diversity index, but the microbial diversity of P. pungens was higher than the two toxic species at this depth when using the Shannon index (Table 2). Although not all possible bacterial OTUs of P. pungens have been sequenced as shown by the unsaturated rarefaction curves for these samples (Figure 1a), the diversity indices at various sequence depths suggest that bacteria associating with the non-toxic species are more diverse than the toxic species (Supplementary Figure S1a and S1b).
Microbial diversities within and between Pseudo-nitzschia samples. (a) Rarefaction curves based on the number of OTUs observed for all Pseudo-nitzschia samples at a sampling depth of 6650 sequences. (b) Clustering of microbial communities as shown by Principal Coordinate Analysis (PCoA) based on Unweighted UniFrac Distance of all samples from three Pseudo-nitzschia species. Orange: P. pungens, red: P. australis, blue: P. fraudulenta.
Furthermore, the microbial communities of the three Pseudo-nitzschia species are significantly different (P⩽0.02) from each other as shown in the principal coordinate analysis based on unweighted UniFrac distance (Figure 1b). The microbial communities within species cluster together and it is notable that the toxic P. fraudulenta has smaller distance to toxic P. australis (UniFrac distance=0.58, P⩽0.02), than to P. pungens (UniFrac distance=0.86, P⩽0.02). These suggest that the microbial communities of the toxic Pseudo-nitzschia are more phylogenetically similar than the non-toxic Pseudo-nitzschia species. The unweighted UniFrac distance of P. australis to P. pungens is 0.87 (P⩽0.02). Consequently, the microbial communities between toxic and non-toxic group are statistically different (UniFrac distance=0.89, P⩽0.0001).
The microbial compositions and the relative abundance of the 16S rDNA sequences of each Pseudo-nitzschia species at the family and genus levels are described and compared in the Supplementary Figures S2 and S3, respectively.
Effect of native microbiota on host fitness
The microbiota isolated from P. pungens and P. australis were re-introduced to their native algal hosts (=source algal species of microbiota), respectively, to determine the influence of each microbiota on host growth rate (i.e., fitness). Remarkably, bacteria-free Pseudo-nitzschia consistently have low fitness (Figures 2 and 3), suggesting that microbiota do indeed have a significant role in the well-being of diatom hosts, just like in animal and plant host–microbiota system (reviewed in Dethlefsen et al., 2007 and Zilber-Rosenberg and Rosenberg, 2008). The SPG rates per day of P. pungens were significantly different when in binary association with its microbiota from Alpha-proteobacteria, Gamma-proteobacteria or Bacteroidetes(F7, 23=20.4, P<0.001) (Figures 2a–c and Supplementary Figure S4a-c). All P. pungens with bacteria have significantly higher SPG than axenic P. pungens.
Algal growth of three Pseudo-nitzschia species co-cultured with individual members of microbiota isolated from P. pungens. (a–c) P. pungens-C5A co-cultured with its own microbiota from Gamma-proteobacteria, Alpha-proteobacteria and Bacteroidetes, respectively. (d–f) P. australis-12 co-cultured with P. pungens microbiota from Gamma-proteobacteria, Alpha-proteobacteria and Bacteroidetes, respectively. (g–i) P. fraudulenta-8 co-cultured with P. pungens microbiota from Gamma-proteobacteria, Alpha-proteobacteria and Bacteroidetes, respectively. Numbers after the name of bacterial treatment in the legend are SPG rates per day of the Pseudo-nitzschia species. Asterisks denote significant differences (P<0.05) between SPG rates of the bacterial treatment and axenic cultures analyzed with post-hoc Student’s t. PP, P. pungens; PA, P. australis; PF, P. fraudulenta; AX, axenic (n=3); ALT, Alteromonas (n=3); MAR, Marinobacter (n=3); PSE, Pseudoalteromonas (n=3); PHAEO, Phaeobacter (n=3); ROSEO, Roseobacter (n=3); CEL, Cellulophaga (n=3); POL, Polaribacter (n=3).
Algal growth of P. australis and P. fraudulenta co-cultured with individual members of microbiota isolated from P. australis. (a–c) P. australis-12 co-cultured with its own microbiota from Gamma-proteobacteria, Alpha-proteobacteria and Bacteroidetes, respectively. (d–f) P. fraudulenta-8 co-cultured with P. australis microbiota from Gamma-proteobacteria, Alpha-proteobacteria and Bacteroidetes, respectively. Numbers after the name of bacterial treatment in the legend are SPG rates per day of the Pseudo-nitzschia species. Asterisks denote significant differences (P<0.05) between SPG rates of the bacterial treatment and axenic cultures analyzed with post-hoc Student’s t-test. PA, P. australis; AX, axenic (n=3); MAR, Marinobacter (n=3); PSE, Pseudoalteromonas (n=3); GLA, Glaciecola (n=3); ROSEO, Roseobacter (n=3); SAL, Salegentibacter (n=3).
P. australis treated with its own bacteria also showed significant differences in SPG (F5, 10=10.4, P<0.001) but these vary between microbiota. When compared with axenic cultures, only the bacteria PA–Marinobacter (Gamma-proteobacteria) and PA–Salegentibacter (Bacteroidetes) significantly enhanced the SPG of their native host, while PA–Roseobacter (Alpha-proteobacteria) significantly retarded its growth (Figures 3a–c). The SPG of P. australis with PA–Pseudoalteromonas and PA–Glaciecola (Gamma-proteobacteria) did not significantly differ from axenic cultures (Figure 3a).
Effect of foreign microbiota on host fitness
The signature of host-specificity and host-adaptation of microbiota can be determined by transplanting the microbiota of one host into another. If the microbiota are not host-specific, the microbiota will display the same influence on the growth of the native and foreign hosts. We transplanted the microbiota of P. pungens into P. australis and P. fraudulenta, and the microbiota of P. australis into P. fraudulenta. We were not able to do a reciprocal transplant of P. australis microbiota to P. pungens because we lost the P. pungens cultures from our collection. Consequently, we expect that the microbiota of P. pungens, which greatly enhances the host growth, will also exhibit the same effect on P. australis and P. fraudulenta. Interestingly, this is not often the case. Responses were either an increase or no effect on the growth of foreign hosts, or the bacteria became algicidal to foreign hosts. For example, P. australis showed significantly different SPG when co-cultured with P. pungens bacteria (F7, 16=9.75, P<0.0001) but this varies with the types of bacteria. P. australis grown with the Gamma-proteobacteria, PP–Marinobacter and PP–Alteromonas showed significantly higher SPG than axenic cultures but not with PP–Pseudoalteromonas (Figure 2d). When co-cultured with the Alpha-proteobacteria from P. pungens, only P. australis co-cultured with PP–Phaeobacter showed an increase in growth rate but these do not significantly differ from P. australis axenic cultures (Figure 2e and Supplementary Figure S4e). Interestingly, PP–Roseobacter, which greatly enhances the growth of its native host, P. pungens, did not have a significant effect on the growth of P. australis (Figure 2e and Supplementary Figure S4e). The Bacteroidetes PP–Polaribacter also significantly increased the SPG of P. australis (Figure 2f and Supplementary Figure S4f). The most remarkable effect is seen with the bacteria PP–Cellulophaga; it significantly promoted the growth of its native host P. pungens but severely depressed the growth of P. australis hosts (Figure 2f and Supplementary Figure S4f).
When bacteria of P. pungens were transplanted to axenic P. fraudulenta, a significant effect of bacteria on algal growth rate is seen (F7,16=10.58, P<0.0001). Mean comparisons of SPG between cultures suggest that only the bacteria PP–Cellulophaga was significantly different against axenic P. fraudulenta cultures (Figures 2g–i). However, algal growth trends showed that the binary cultures with the other six bacteria crashed earlier than the axenic cultures at day 8.
The transplant experiment of P. australis microbiota to P. fraudulenta showed no effect on SPG of the foreign host (F5, 12=2.44, P=0.09) (Figure 3). PA–Roseobacter, which severely retarded the growth of P. australis (Figure 3b), did not exhibit any of these effects on P. fraudulenta (Figure 3e). Notably, PA–Marinobacter and PA–Salegentibacter, which can both increase the growth rate of P. australis, did not show such effects on P. fraudulenta (Figures 3d and f).
Response of non-axenic Pseudo-nitzschia to an algicidal bacterium
We further investigated the interaction of PP–Cellulophaga that is algicidal to axenic P. australis and P. fraudulenta but mutualistic to P. pungens. We supposed that the two foreign hosts are weakened when in the axenic state and were vulnerable to algalytic attack but not when associating with bacteria. We therefore introduced PP–Cellulophaga to other three non-axenic strains of P. australis and P. fraudulenta to determine their susceptibility to PP–Cellulophaga. Again we found contrasting responses between the two foreign hosts. We found that the SPG of non-axenic P. fraudulenta clones were consistently not affected by the presence of Cellulophaga (PF-1: F1, 4=0.53, P=0.51; PF-4: F1, 4=3.74, P=0.13; PF-8: F1, 4=3.15, P=0.15) (Figures 4a–c). The non-axenic three strains of P. australis treated with PP–Cellulophaga showed consistent lower growth rates, but only the SPG of PA-15 showed statistical difference against its untreated counterpart (PA-10: F1, 4=4.99, P=0.09; PA-12: F1, 4=3.03, P=0.16; PA-15: F1, 4=49.2, P=0.002) (Figures 4d–f).
Varying growth responses of three non-axenic (NON-AX-with normal microflora) P. fraudulenta and P. australis clones when challenged with an algicidal bacterium, PP–CEL (Cellulophaga). (a–c) P. fraudulenta (PF) clone numbers 1, 4 and 8. (d–f) P. australis (PA) clone numbers 10, 12, 15. Solid lines: Pseudo-nitzschia cultures with normal microflora only; dashed lines: Pseudo-nitzschia cultures with normal microflora and grown with Cellulophaga bacterium. Numbers after the name of bacterial treatment in the legend are SPG rates per day of the Pseudo-nitzschia species. Asterisks denote significant differences (P<0.05) between SPG rates of the bacterial treatment and axenic cultures analyzed with one-way analysis of variance (ANOVA). Each treatment has n=3 replicates.
Influence of single bacterium on DA production of Pseudo-nitzschia
We investigated the effects of different bacteria on the production of DA of toxic Pseudo-nitzschia species. We expressed the concentration of DA both as cellular DA (femtogram per cell; Figure 5) and particulate DA (μg l−1; Supplementary Figure S5). In general, we found that foreign microbiota enhances production of DA of the toxigenic Pseudo-nitzschia. P. australis DA production range from 38.2–1398.5 femtogram per cell (or 44.7–823.7 μg l−1) depending on the type of microbiota it harbors. The overall mean DA content of all P. australis cultures is 312 femtogram per cell (or 280.5 μg l−1). Mean comparisons showed that P. australis grown with P. pungens microbiota (PP–Alteromonas and PP–Glaciecola, and PP–Polaribacter ) have statistically higher DA content than the overall mean (2.5–5 times higher, P<0.05), and 3–7 times more DA compared with axenic P. australis (Figure 5a and Supplementary Figure S5a). P. australis treated with the Alpha-proteobacteria, PP–Roseobacter, has statistically lower DA content than overall mean (8 times lower, P<0.05), 5 times less DA than the axenic cultures. Interestingly, all five microbiota members of P. australis in this study and the Firmicutes PP–Bacillus and PP–/PA–Planococcus did not statististically differ from the overall mean DA production of P. australis (Figure 5a and Supplementary Figure S5a). We also introduced the native microbiota of P. pungens to the same P. pungens host clones, but no bacteria induced DA production in the non-toxic P. pungens (data not shown).
The effects of different bacteria on DA production of P. fraudulenta, on the other hand, showed a different pattern. P. fraudulenta does not produce a high amount of DA as compared with P. australis, ranging from 0–30.7 femtogram per cell or (0–25 μg l−1). Its overall mean DA content is 6.5 femtogram per cell (or 10.1 μg l−1). Interestingly, some microbiota of P. australis (PA–Salegentibacter, PA–Marinobacter and PA–Planococcus) significantly enhanced its cellular DA production (3–5 times higher than overall mean, P<0.05) (Figure 5b and Supplementary Figure S5b). Both Bacteroidetes microbiota from P. pungens (PP–Cellulophaga and PP–Polaribacter) also significantly increased its DA content by 1.5–1.8 times (Figure 5 and Supplementary Figure S5b). Notably, four P. pungens microbiota (PP–Roseobacter, PP–Phaeobacter, PP–Alteromonas and PP–Bacillus) and the P. australis microbiota, PA–Glaciecola, rendered P. fraudulenta non-toxic (<1 μg l−1 or femtogram per cell). DA contents of P. fraudulenta with these five bacteria were all significantly lower than the overall mean. It is also interesting to note that axenic P. fraudulenta has significantly higher DA content than the overall mean (two times higher, P<0.05) and is both higher than the unaltered microbiota (NON-AX) of P. australis and P. fraudulenta, which is in contrary to reports of Bates et al. (1995) and Douglas et al. (1993). Our results, however, are consistent with Stewart (2008) which suggest that bacteria putatively consume the DA produced by its algal host. Axenic condition may also be stressful for the algae, hence, the production of higher DA than non-axenic cultures.
Discussion
This study is the first to implement an evolutionary perspective in understanding the interaction between phytoplankton and bacterial associates. Our study showed that the marine diatom Pseudo-nitzschia not only associates with a distinct and diverse group of bacteria but also governs the relationships through host-specificity and co-adaptation. The natural partners confer better host fitness than the artificially combined partners, which hints towards co-evolution of Pseudo-nitzschia and their microbiota. This is best exemplified by the mutualistic interactions of the parasitic bacteria PP–Cellulophaga and the commensal PP–Roseobacter to their host P. pungens. Such adaptation is evidenced by the low degree of virulence of these bacteria towards their own host but algalytic or commensal to non-hosts. Marine bacteria are known to switch on and off their algicidal activity through enzyme production (Skerratt et al., 2002), which may explain our observation in a host-dependent manner. In retrospect, the Pseudo-nitzschia hosts are adapted to their native microbiota as they exhibited positive growth even in the presence of microbiota with algicidal and commensal traits, a suggestion of co-adaptation between partners.
The requirement for the evolution of mutualistic relationship between distantly related organisms is constant proximity (Usher et al., 2007), such as that seen in diatoms–cyanobacteria associations (Janson 2003; Usher et al., 2007). Recognition between partners, therefore, is necessary to maintain such relationship, likely in the form of chemical communication. We hypothesized that the type of algal exudate secretions by the hosts maybe regulating the mutualistic or parasitic relationship between bacteria and diatom hosts. Indeed, algal hosts are quite distinct in their excretions of organic molecules (Hellebust, 1980; Myklestad, 1995), and different forms of excretions (e.g., polysaccharides) can be utilized by different bacterial strains (Bruckner et al., 2011). Studying the algal exudate excretions of different Pseudo-nitzschia species and measuring the bacterial enzyme secretions when in binary association with a microbiota that exhibit both mutualistic and parasitic traits would shed light into the evolution and regulation of these complex relationships.
In the light of our results, adaptation and specificity between bacteria and phytoplankton hosts may have an implication on the ecological structuring of phytoplankton communities in the ocean. For instance, the microbiota of one algal species may become an agent of allelopathic means during competition, where a neighboring algal species can be negatively affected once invaded by the foreign bacteria. Accordingly, our study also showed that a microbiota could ameliorate the susceptibility of phytoplankton to parasitic bacteria, as in the case of non-axenic P. fraudulenta when challenged with the algalytic PP–Cellulophaga (Figure 4a). This may perhaps be another evolutionary strategy of phytoplankton in counteracting parasitic bacteria, although this may not be the case with P. australis. But it is noteworthy to mention that P. australis already harbors parasitic bacteria (PA–Roseobacter) in its microbiota consortium which may have increased its susceptibility to parasitic attack. Unlike metazoans, marine diatoms are not known to have an immune system that can counteract parasites. Duff et al. (1966) suggested that marine diatoms can secrete antibiotics but this has not been verified by other studies. Previous reports hypothesized that DA is an anti-bacterial agent (Bates et al., 1995). This does not seem to be the case with the toxigenic Pseudo-nitzschia, because the diatom still remained susceptible to parasitic bacteria and did not show any elevation of DA secretion when challenged with parasitic bacteria (Figure 5). Others have also reported that epibiont bacteria must be present for Pseudo-nitzschia to undergo sexual reproduction (Thompson, 2000 as reported by Trainer et al. (2012)), suggesting that DA is unlikely to be a non-selective anti-bacterial agent. It is interesting to note the functional similarity of microbiota with regards to conferring protection to its hosts, whether it is with the unicellular host (this study) or with the vertebrate hosts (Kosiewicz et al., 2011).
We asked if algal species could influence the microbial community structure of Pseudo-nitzschia. Indeed, the microbial communities of the three species of Pseudo-nitzschia are significantly distinct from each other, and the microbial communities of the toxic species are much more closely related to each other than the non-toxic species. This host-defined structuring of microbial communities is consistent with other reports that looked at bacterial communities of different diatoms in culture (Grossart et al., 2005; Sapp et al., 2007; Guannel et al., 2011). Two possibilities can bring about this observed host-specific structure in microbial communities. One possibility is that these could be the effect of temporal and biogeographical variation in the algal hosts collection. However, this is not the case in our samples, because we collected them at the same place, although there is a 3–6 month time difference between collections. An independent study, however, that is consistent with our results, showed that Pseudo-nitzschia species have microbial communities that are species-specific (Guannel et al., 2011) even when the algae were collected at different times and biogeographic regions. An alternative possibility is that algal hosts are quite distinct in their secretions of organic molecules (Hellebust, 1980; Myklestad, 1995), which can restrict bacteria in the community assembly. In a freshwater system, for example, bacterial communities that have the genetic potential to utilize glycolate are structured accordingly by the shift in phytoplankton assemblage that releases this exudate (Paver et al., 2010). The three species of Pseudo-nitzschia in this study have a varying degree of DA production and have been found to harbor distinct microbial communities. In addition, the non-toxic P. pungens associates with a much more diverse bacterial assemblage than toxic P. australis and P. fraudulenta, a good indication that DA can be a restricting factor in the assembly of bacterial communities.
Many factors can bring about the phenotypic plasticity in DA production of Pseudo-nitzschia (see review by Trainer et al., 2012). With regards to bacterial influence, Bates et al., (1995) first showed that foreign bacteria could enhance the DA production of axenic P. multiseries, while Stewart (2008) suggest that co-occurring bacteria in Pseudo-nitzschia cultures may consume DA. In our study, we asked which bacteria assume these roles. Interestingly, only three bacteria (two Gamma-proteobacteria and one Bacteroidetes) from P. pungens elevated the DA production of P. australis, while its own microbiota did not. The DA productions of P. fraudulenta, on the other hand, were enhanced by P. australis microbiota, while DA is barely measurable when treated with P. pungens Alpha-and Gamma-proteobacteria microbiota. In general, only foreign bacteria seem to trigger the elevated production of DA, which is viewed previously as a defense mechanism of toxic Pseudo-nitzschia (Bates et al., 1995; Kaczmarska et al., 2005) against foreign bacteria. Our study rather suggests that elevated DA is a mere stress response to these bacteria, just like any other environmental stresses experienced by toxigenic Pseudo-nitzschia that causes the organism to produce elevated DA (see review by Trainer et al. (2012)). Indeed, one pattern observed in our study was that most foreign bacteria were mostly commensals and had no obvious effect on the growth of non-hosts diatom. It remains to be seen if these patterns holds true when P. australis or P. fraudulenta is co-cultured with the microbiota of other Pseudo-nitzschia species.
The type of interactions we observed between Pseudo-nitzschia and their microbiota in this study are confined to uniform laboratory conditions and only binary interactions have been characterized, which are simplified conditions designed to tease apart the complex interactions of diatom–bacteria relationship. Other interacting environmental factors such as pH, temperature or nutrient limitation can also shape algal–bacterial interactions, and may possibly deviate from the patterns we observed in our study. Consequently, our culture medium is meant to simulate a condition for algal bloom, where Pseudo-nitzschia can thrive in dense numbers, and have quantifiable toxin production, thus, making this work relevant to algal bloom studies. Studying the interactions under these conditions provide an insight into the patterns that govern the dynamic and complex relationship of algae and their associated bacteria and also shed light into the influence of microbiota in the formation of algal bloom (and its demise) and variability of toxin production in Pseudo-nitzschia. The Pseudo-nitzschia-bacteria system is also amenable to experimental manipulations and can be used to dissect the role of associated bacteria on host responses to biotic changes (for example, the presence of parasitic bacteria in this study). The genus Pseudo-nitzschia is also one of the most studied diatoms in terms of biology, ecology and physiology and with an available genome and transcriptomes, which makes it an ideal model system for studying bacteria–phytoplankton interactions and relating the findings in a biological and ecological framework.
In summary, the combination of pyrosequencing, fitness assay and physiological measurement have demonstrated that the distinct and diverse bacterial consortium of the marine diatom, Pseudo-nitzschia, generally are mutualist to the native host and commensal or parasitic to foreign hosts. These interactions clearly suggest that host-specificity and co-adaptation are involved in the relationship. Harboring microbiota may also give protection to the algal host from pathogenic bacteria and may be a means of allelopathy by negatively affecting the fitness of other algal species. This study also demonstrates that algal species and its metabolite production may have a role in structuring the microbial communities of a unicellular eukaryotic host. Different bacteria can also have varying influence on the DA production of toxic Pseudo-nitzschia, and can alter the diatom’s presumed toxicity, a knowledge that should be taken into consideration in the management of Pseudo-nitzschia bloom.
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ . (1990). Basic local alignment search tool. J Mol Biol 215: 403–410.
Amin SA, Green DH, Hart MC, Kupper FC, Sunda WG, Carrano CJ . (2009). Photolysis of iron-siderophore chelates promotes bacterial-algal mutualism. Proc Natl Acad Sci USA 106: 17071–17076.
Armbrust EV . (2009). The life of diatoms in the world's oceans. Nature 459: 185–192.
Armbrust EV, Berges JA, Bowler C, Green BR, Martinez D, Putnam NH et al (2004). The genome of the diatom Thalassiosira pseudonana: Ecology, evolution, and metabolism. Science 306: 79–86.
Azam F . (1998). Microbial control of oceanic carbon flux: The plot thickens. Science 280: 694–696.
Azam F, Fenchel T, Field JG, Gray JS, Meyerreil LA, Thingstad F . (1983). The ecological role of water-column microbes in the sea. Mar Ecol Prog Ser 10: 257–263.
Bates SS, Bird CJ, De Freitas ASW, Foxall R, Gilgan M, Hanic LA et al (1989). Pennate diatom Nitzschia- pungens as the primary source of domoic acid, a toxin in shellfish from Eastern Prince Edward Island, Canada. Can J Fish Aquat Sci 46: 1203–1215.
Bates SS, Douglas DJ, Doucette GJ, Leger C . (1995). Enhancement of domoic acid production by reintroducing bacteria to axenic cultures of the diatom Pseudo-nitzschia multiseries. Nat Toxins 3: 428–435.
Bell W, Mitchell R . (1972). Chemotactic and growth responses of marine bacteria to algal extracellular products. Biol Bull 143: 265–277.
Bowler C, Allen AE, Badger JH, Grimwood J, Jabbari K, Kuo A et al (2008). The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456: 239–244.
Bruckner CG, Bahulikar R, Rahalkar M, Schink B, Kroth PG . (2008). Bacteria associated with benthic diatoms from Lake Constance: Phylogeny and influences on diatom growth and secretion of extracellular polymeric substances. Appl Environ Microbiol 74: 7740–7749.
Bruckner CG, Rehm C, Grossart H-P, Kroth PG . (2011). Growth and release of extracellular organic compounds by benthic diatoms depend on interactions with bacteria. Environ Microbiol 13: 1052–1063.
Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R . (2010). PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26: 266–267.
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al (2010). QIIME allows analysis of high-throughput community sequencing data. Nat Meth 7: 335–336.
Cole JJ . (1982). Interactions between bacteria and algae in aquatic ecosystems. Annu Rev Ecol Syst 13: 291–314.
DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K et al (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72: 5069–5072.
Dethlefsen L, McFall-Ngai M, Relman DA . (2007). An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature 449: 811–818.
Doucette GJ . (1995). Interactions between bacteria and harmful algae: a review. Nat Toxins 3: 65–74.
Doucette GJ, Kodama M, Franca S, Gallacher S . (1998). Bacterial interactions with harmful algal bloom species: bloom ecology, toxigenesis, and cytology. In: Anderson DM, Cembella AD, Hallegraeff GM, (eds) Physiological Ecology of Harmful Algal Blooms. NATO ASI Series 41. Springer: Berlin, Heidelberg, New York, pp 619–647.
Douglas DJ, Bates SS, Bourque LA, Selvin RC . (1993). Domoic acid production by axenic and non-axenic cultures of the pennate diatom Nitzschia-pungens f. multiseries. In: Smayda TJ, Shimizu Y, (eds) Toxic Phytoplankton Blooms in the Sea. Develop Mar Biol 3. Elsevier Science Publishers: NewYork, NY, USA, pp 595–600.
Dowd SE, Callaway TR, Wolcott RD, Sun Y, McKeehan T, Hagevoort RG et al (2008). Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP). BMC Microbiol 8: 125.
Duff DC, Bruce DL, Antia NJ . (1966). The antibacterial activity of marine planktonic algae. Can J Microbiol 12: 877–884.
Dziallas C, Grossart H-P . (2011a). Temperature and biotic factors influence bacterial communities associated with the cyanobacterium Microcystis sp. Environ. Microbiol 13: 1632–1641.
Dziallas C, Grossart H-P . (2011b). Increasing oxygen radicals and water temperature select for toxic Microcystis sp. PLoS One 6: 9.
Ebert D, Qi W . (2010). An evolutionary ecology perspective on comparative metagenomics. In: Metagenomics and its Applications in Agriculture, Biomedicine, and Environmental Studies, 211–236.
Edgar RC . (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26: 2460–2461.
Falkowski PG, Barber RT, Smetacek V . (1998). Biogeochemical controls and feedbacks on ocean primary production. Science 281: 200–206.
Ferrier M, Martin JL, Rooney-Varga JN . (2002). Stimulation of alexandrium fundyense growth by bacterial assemblages from the Bay of Fundy. J Appl Microbiol 92: 706–716.
Gärdes A, Iversen MH, Grossart H-P, Passow U, Ullrich MS . (2010). Diatom-associated bacteria are required for aggregation of Thalassiosira weissflogii. ISME J 5: 436–445.
Gontcharova V, Youn E, Wolcott RD, Hollister EB, Gentry TJ, Dowd SE . (2010). Black Box Chimera Check (B2C2): a windows-based software for batch depletion of chimeras from bacterial 16S rRNA gene datasets. Open Microbiol J 4: 47–52.
Grossart HP . (1999). Interactions between marine bacteria and axenic diatoms (Cylindrotheca fusiformis, Nitzschia laevis, and Thalassiosira weissflogii) incubated under various conditions in the lab. Aquat Microb Ecol 19: 1–11.
Grossart HP, Levold F, Allgaier M, Simon M, Brinkhoff T . (2005). Marine diatom species harbour distinct bacterial communities. Environ Microbiol 7: 860–873.
Guannel ML, Horner-Devine MC, Rocap G . (2011). Bacterial community composition differs with species and toxigenicity of the diatom Pseudo-nitzschia. Aquat Microb Ecol 64: 117–133.
Guillard RRL, Ryther JH . (1962). Studies of marine planktonic diatoms. I. Cyclotella nana Hustedt and Detonula confervacea Cleve. Can J Microbiol 8: 229–239.
Hellebust JA . (1980). Citation classic—excretion of some organic-compounds by marine-phytoplankton. Curr Contents Agric Biol Environ Sci, 16.
Hoagland P, Anderson DM, Kaoru Y, White AW . (2002). The economic effects of harmful algal blooms in the United States: Estimates, assessment issues, and information needs. Estuaries 25: 819–837.
Hoben HJ, Somasegaran P . (1982). Comparison of the pour, spread, and drop plate methods for enumeration of Rhizobium spp in inoculants made from pre-sterilized peat. Appl Environ Microbiol 44: 1246–1247.
Huse SM, Huber JA, Morrison HG, Sogin ML, Mark Welch D . (2007). Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol 8: R143.
Janson S . (2003). Cyanobacteria in symbiosis with diatoms. in Rai AN, Bergman B, Rasmussen U, (eds) Cyanobacteria in Symbiosis, 1–10 Kluwer Academic Publishers: The Netherlands.
Kaczmarska I, Ehrman JM, Bates SS, Green DH, Leger C, Harris J . (2005). Diversity and distribution of epibiotic bacteria on Pseudo-nitzschia multiseries (Bacillariophyceae) in culture, and comparison with those on diatoms in native seawater. Harmful Algae 4: 725–741.
Kosiewicz MM, Zirnheld AL, Alard P . (2011). Gut microbiota, immunity, and disease: a complex relationship. Frontiers Microbiol 2: 180.
Litaker RW, Stewart TN, Eberhart BTL, Wekell JC, Trainer VL, Kudela RM et al (2008). Rapid enzyme-linked immunosorbent assay for Detection of the algal toxin domoic acid. J Shellfish Res 27: 1301–1310.
Liu JQ, Lewitus AJ, Brown P, Wilde SB . (2008). Growth-promoting effects of a bacterium on raphidophytes and other phytoplankton. Harmful Algae 7: 1–10.
Mayali X, Azam F . (2004). Algicidal bacteria in the sea and their impact on algal blooms. J Eukaryot Microbiol 51: 139–144.
Mouget JL, Dakhama A, Lavoie MC, Delanoue J . (1995). Algal growth enhancement by bacteria—is consumption of photosynthetic oxygen involved. FEMS Microbiol Ecol 18: 35–43.
Myklestad SM . (1995). Release of extracellular products by phytoplankton with special emphasis on polysaccharides. Sci Total Environ 165: 155–164.
Paver SF, Kent AD . (2010). Temporal patterns in glycolate-utilizing bacterial community composition correlate with phytoplankton population dynamics in humic lakes. Microb Ecol 60: 406–418.
Perl TM, Bédard L, Kosatsky T, Hockin JC, Todd EC, Remis RS . (1990). An outbreak of toxic encephalopathy caused by eating mussels contaminated with domoic acid. N Engl J Med 322: 1775–1780.
Price MN, Dehal PS, Arkin AP . (2010). FastTree 2-approximately maximum-likelihood trees for large alignments. PLoS One 5: e9490.
Sapp M, Schwaderer AS, Wiltshire KH, Hoppe HG, Gerdts G, Wichels A . (2007). Species-specific bacterial communities in the phycosphere of microalgae? Microb Ecol 53: 683–699.
Skerratt JH, Bowman JP, Hallegraeff G, James S, Nichols PD . (2002). Algicidal bacteria associated with blooms of a toxic dinoflagellate in a temperate Australian estuary. Marine Ecol 244: 1–15.
Stewart JE . (2008). Bacterial involvement in determining domoic acid levels in Pseudo-nitzschia multiseries cultures. Aquat Microb Ecol 50: 135–144.
Stewart JE, Marks LJ, Wood CR, Risser SM, Gray S . (1997). Symbiotic relations between bacteria and the domoic acid producing diatom Pseudo-nitzschia multiseries and the capacity of these bacteria for gluconic acid gluconolactone formation. Aquat Microb Ecol 12: 211–221.
Sun Y, Wolcott RD, Dowd SE . (2011). Tag-encoded FLX amplicon pyrosequencing for the elucidation of microbial and functional gene diversity in any environment. High-Throughput Next Generation Sequen Methods Appl 733: 129–141.
Thompson S . (2000). The role of bacteria in mediating the sexual reproduction of the domoic-acid-producing diatom Pseudo-nitzschia multiseries (Hasle) Hasle. Special Topics Thesis. Mount Allison University: Sackville, NB, 49.
Trainer VL, Bates SS, Lundholm N, Thessen AE, Cochlan WP, Adams NG et al (2012). Pseudo-nitzschia physiological ecology, phylogeny, toxicity, monitoring and impacts on ecosystem health. Harmful Algae 14: 271–300.
Trainer VL, Cochlan WP, Erickson A, Bill BD, Cox FH, Borchert JA et al (2007). Recent domoic acid closures of shellfish harvest areas in Washington State inland waterways. Harmful Algae 6: 449–459.
Usher KM, Bergman B, Raven JA . (2007). Exploring cyanobacterial mutualisms. Annu Rev Ecol Evol Syst 38: 255–273.
Wang Q, Garrity GM, Tiedje JM, Cole JR . (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73: 5261–5267.
Wells ML, Trick CG, Cochlan WP, Hughes MP, Trainer VL . (2005). Domoic acid: The synergy of iron, copper, and the toxicity of diatoms. Limnol Oceanogr 50: 1908–1917.
Zilber-Rosenberg I, Rosenberg E . (2008). Role of microorganisms in the evolution of animals and plants: the hologenome theory of evolution. FEMS Microbiol Rev 32: 723–735.
Acknowledgements
We gratefully thank Kendra Hayashi for providing plankton tow samples and Lucy Li for helping with bacterial cultures. The National Science Foundation OCE 1131770 supported this work. UC Presidential Postdoctoral Fellowship supported MPSM. Undergraduate Research Opportunities Program—UC Irvine, funded KNT. Collection of environmental data and Pseudo-nitzschia isolates from the Santa Cruz Wharf was supported by Cal-PReEmpt with funding from the NOAA-MERHAB program (Grant no. NA04NOS4780239 MERHAB publication #175).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Sison-Mangus, M., Jiang, S., Tran, K. et al. Host-specific adaptation governs the interaction of the marine diatom, Pseudo-nitzschia and their microbiota. ISME J 8, 63–76 (2014). https://doi.org/10.1038/ismej.2013.138
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2013.138
Keywords
This article is cited by
-
Algal blooms in the ocean: hot spots for chemically mediated microbial interactions
Nature Reviews Microbiology (2024)
-
Degradation of polycyclic aromatic hydrocarbons in aquatic environments by a symbiotic system consisting of algae and bacteria: green and sustainable technology
Archives of Microbiology (2024)
-
Host specificity of microbiome assembly and its fitness effects in phytoplankton
The ISME Journal (2021)
-
A novel random forest approach to revealing interactions and controls on chlorophyll concentration and bacterial communities during coastal phytoplankton blooms
Scientific Reports (2021)
-
Genomic insights into the molecular mechanisms of a Pseudomonas strain significant in its survival in Kongsfjorden, an Arctic fjord
Molecular Genetics and Genomics (2021)