Abstract
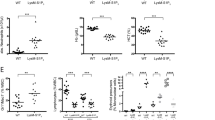
Constitutive activation of the PI3K/AKT signaling pathway is found in ~50–70% of AML patients. The SH2-containing inositol 5-phosphatase 1 (SHIP1) is a negative regulator of PI3K/AKT signaling in hematopoietic cells. SHIP1 knockout mice develop a myeloproliferative syndrome and concomitant deletion of SHIP1 and the tumor suppressor PTEN leads to the development of lethal B-cell lymphomas. In the study presented here, we investigated the role of SHIP1 as a tumor suppressor in myeloid leukemia cells in an in vivo xenograft transplantation model. NSG Mice transplanted with UKE-1 cells derived from a secondary AML showed a significantly extended lifespan after lentiviral-mediated overexpression of SHIP1 in comparison to the vector control cohort. In contrast, the AML-derived SHIP1Y643H mutant, which has a strongly reduced enzymatic activity showed a significant reversion of the SHIP1-induced prolongation of the survival time. In addition, the analysis of 290 AML patients revealed a correlation between expression of SHIP1 and overall survival of the AML patients. These results indicate that SHIP1 can act as a tumor suppressor in acute myeloid leukemia cells and that higher SHIP1 expression is associated with prolonged overall survival in AML patients. SHIP1 may be an interesting candidate for gene therapy.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Accession codes
References
Martelli AM, Evangelisti C, Chiarini F, McCubrey JA . The phosphatidylinositol 3-kinase/Akt/mTOR signaling network as a therapeutic target in acute myelogenous leukemia patients. Oncotarget 2010; 1: 89–103.
Helgason CD, Damen JE, Rosten P, Grewal R, Sorensen P, Chappel SM et al. Targeted disruption of SHIP leads to hemopoietic perturbations, lung pathology, and a shortened life span. Genes Dev 1998; 12: 1610–1620.
Liu Q, Sasaki T, Kozieradzki I, Wakeham A, Itie A, Dumont DJ et al. SHIP is a negative regulator of growth factor receptor-mediated PKB/Akt activation and myeloid cell survival. Genes Dev 1999; 13: 786–791.
Miletic AV, Anzelon-Mills AN, Mills DM, Omori SA, Pedersen IM, Shin DM et al. Coordinate suppression of B cell lymphoma by PTEN and SHIP phosphatases. J Exp Med 2010; 207: 2407–2420.
O'Connell RM, Chaudhuri AA, Rao DS, Baltimore D . Inositol phosphatase SHIP1 is a primary target of miR-155. Proc Natl Acad Sci USA 2009; 106: 7113–7118.
Pedersen IM, Otero D, Kao E, Miletic AV, Hother C, Ralfkiaer E et al. Onco‐miR‐155 targets SHIP1 to promote TNFα‐dependent growth of B cell lymphomas. EMBO Mol Med 2009; 1: 288–295.
Xue H, Hua LM, Guo M, Luo JM . SHIP1 is targeted by miR-155 in acute myeloid leukemia. Oncol Rep 2014; 32: 2253–2259.
Metzner A, Precht C, Fehse B, Fiedler W, Stocking C, Gunther A et al. Reduced proliferation of CD34(+) cells from patients with acute myeloid leukemia after gene transfer of INPP5D. Gene Ther 2009; 16: 570–573.
Metzner A, Horstmann MA, Fehse B, Ortmeyer G, Niemeyer CM, Stocking C et al. Gene transfer of SHIP-1 inhibits proliferation of juvenile myelomonocytic leukemia cells carrying KRAS2 or PTPN11 mutations. Gene Ther 2007; 14: 699–703.
Horn S, Endl E, Fehse B, Weck MM, Mayr GW, Jucker M . Restoration of SHIP activity in a human leukemia cell line downregulates constitutively activated phosphatidylinositol 3-kinase/Akt/GSK-3beta signaling and leads to an increased transit time through the G1 phase of the cell cycle. Leukemia 2004; 18: 1839–1849.
Brauer H, Strauss J, Wegner W, Muller-Tidow C, Horstmann M, Jucker M . Leukemia-associated mutations in SHIP1 inhibit its enzymatic activity, interaction with the GM-CSF receptor and Grb2, and its ability to inactivate PI3K/AKT signaling. Cell Signal 2012; 24: 2095–2101.
James C, Ugo V, Le Couedic JP, Staerk J, Delhommeau F, Lacout C et al. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature 2005; 434: 1144–1148.
Quentmeier H, MacLeod RA, Zaborski M, Drexler HG . JAK2 V617F tyrosine kinase mutation in cell lines derived from myeloproliferative disorders. Leukemia 2006; 20: 471–476.
Puissant A, Fenouille N, Alexe G, Pikman Y, Bassil CF, Mehta S et al. SYK is a critical regulator of FLT3 in acute myeloid leukemia. Cancer Cell 2014; 25: 226–242.
Verhaak RG, Wouters BJ, Erpelinck CA, Abbas S, Beverloo HB, Lugthart S et al. Prediction of molecular subtypes in acute myeloid leukemia based on gene expression profiling. Haematologica 2009; 94: 131–134.
Chen Z, Shojaee S, Buchner M, Geng H, Lee JW, Klemm L et al. Signalling thresholds and negative B-cell selection in acute lymphoblastic leukaemia. Nature 2015; 521: 357–361.
Fiedler W, Henke RP, Ergun S, Schumacher U, Gehling UM, Vohwinkel G et al. Derivation of a new hematopoietic cell line with endothelial features from a patient with transformed myeloproliferative syndrome: a case report. Cancer 2000; 88: 344–351.
Weber K, Mock U, Petrowitz B, Bartsch U, Fehse B . Lentiviral gene ontology (LeGO) vectors equipped with novel drug-selectable fluorescent proteins: new building blocks for cell marking and multi-gene analysis. Gene Ther 2010; 17: 511–520.
Acknowledgements
We acknowledge with special thanks the excellent technical assistance of Bettina Bettin, Christine Blechner und Susanne Giehler. This study was supported by a post-doctoral fellowship from the Werner Otto Stiftung, Hamburg, Germany to MT, a grant from the Hamburger Krebsgesellschaft to SH and a grant from the Erich und Gertrud Roggenbuck-Stiftung to MJ.
Author contributions
MT designed and performed experiments, analyzed results and wrote the manuscript; SH and JW designed and performed experiments, analyzed results; EL assisted mouse experiments; JW performed bioinformatics analysis; PE and MS performed western blot and RT-PCR analysis; MN and BF provided the material and gave advice; WF and CS contributed to data collection and approved the final version of the manuscript; MJ designed the research, analyzed data and wrote the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on Gene Therapy website
Supplementary information
Rights and permissions
About this article
Cite this article
Täger, M., Horn, S., Latuske, E. et al. SHIP1, but not an AML-derived SHIP1 mutant, suppresses myeloid leukemia growth in a xenotransplantation mouse model. Gene Ther 24, 749–753 (2017). https://doi.org/10.1038/gt.2017.88
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gt.2017.88
This article is cited by
-
Targeted PI3K/AKT-hyperactivation induces cell death in chronic lymphocytic leukemia
Nature Communications (2021)