Abstract
Newcastle disease virus (NDV) is an oncolytic paramyxovirus with a nonsegmented single-stranded RNA genome. In this report, a recombinant oncolytic NDV was passaged in human tumor xenografts and reisolated and characterized after two rounds of bioselection. Several isolates could be recovered that differed from the parental virus with respect to virus spread in tumor cells and the ability to form syncytia in human tumor cells. Three isolates were identified that demonstrated superior oncolytic potency compared with the parental virus as measured by increased oncolytic potency in confluent tumor cell monolayers, in tumor cell spheroids and in a mouse xenograft tumor model. The surface proteins F and HN were sequence analyzed and characterized for fusogenicity. The present study demonstrates that in vivo NDV bioselection can enable the isolation of novel, oncolytic NDV and thus represents a powerful methodology for the development of highly potent oncolytic viruses.
Similar content being viewed by others
Introduction
Newcastle disease virus (NDV) is a naturally occurring oncolytic virus that inherently replicates in most human tumor cells. The mechanism of the tumor-selective replication is only partially understood. It is thought that a defect in the cellular interferon response, which is a common observation in human tumor cells, contributes largely to that effect.1 Because of the defective interferon response of tumor cells, selective replication and associated cytotoxicity occurs only in tumor cells, whereas sparing normal cells.2 It has been proposed that apoptosis is involved in the virus-mediated cell killing.3 Unlike other oncolytic viruses, NDV is not dependent on a receptor that is specifically expressed on tumor cells. In contrast, the virus binds to the ubiquitous sialic acid present in glycoproteins on the cell surface. Thus, NDV may be a broad-acting oncolytic agent.
NDV is not pathogenic for humans but it is a well-studied pathogen for birds, in particular chickens.4 Various strains of NDV have been tested for the treatment of human tumors (history reviewed in5 and some of the strains have now been tested in clinical trials.6, 7). The NDV strains are generally classified into three groups (lentogenic, mesogenic and velogenic) based on their pathogenicity in chickens.
Lentogenic viruses are generally considered vaccines, whereas mesogenic and velogenic viruses are generally considered pathogens.4 Because lentogenic NDVs can infect human tumor cells but normally cannot replicate in the tumor cells, they lack the self-amplification property that is a main characteristic of the mesogenic NDVs that are therapeutic candidates. Therefore, clinical trials with oncolytic NDV strains to date have generally been conducted with mesogenic strains. However, there is also a lentogenic strain being tested for tumor treatment.8 Consequently, when engineering the NDV genome or selecting for genetic variants that have a higher oncolytic potency and might be advanced towards clinical testing, it is necessary to minimize the environmental risk to prevent the inadvertant generation of more pathogenic viruses for the natural host, especially poultry.
Several NDV strains have been genetically engineered to alter selected properties, starting with vaccine strains9, 10, 11 and later extending to oncolytic strains.12, 13 Pühler et al.12 describe the generation of a recombinant virus from the naturally oncolytic mesogenic NDV strain, MTH68, expressing the two chains of a full IgG antibody as a transgene. Furthermore, the transgene green fluorescent protein (GFP) was engineered into the viral genome to create an oncolytic reporter virus that can be monitored for the infection of human tumor cells both in vitro and in vivo. Currently, both natural and genetically modified NDV viruses are being studied as oncolytic viruses for virotherapy of tumors.
In monolayers of human tumor cells, one NDV virus particle is sufficient to kill more than 107 cells. However, the 100% cytotoxicity that is often observed in vitro does not translate generally to complete responses in human tumor xenograft experiments (Beier et al., unpublished observations). Further studies indicated that intratumoral virus spread is limited in the xenograft tissue due to barriers that impede the oncolysis in a tumor as an organ as compared with monolayer tissue culture cells. Methods are thus needed that develop viruses that can overcome these limiting barriers in vivo. In an elegant approach to develop highly potent viruses, Kuhn et al.14 described and validated an in vitro methodology for the generation of more potent oncolytic adenoviruses by directed evolution. They forced recombination between various strains of adenoviruses to generate a novel, highly potent oncolytic virus, ColoAd1. This method, however, is limited to viruses with double-stranded genomes. For single-stranded negative genome viruses, such as paramyxoviruses, recombination is not expected to occur and bioselection would only be possible on the basis of stochastically occurring single mutations. Consequently, new methods are needed to exploit the power of bioselection for this class of virus. In this manuscript, we describe and validate that bioselection of oncolytic NDV in vivo in human tumor xenografts is a powerful method to isolate viruses that have higher oncolytic potency than their parental strain both in vitro and in vivo.
Results
Bioselection of NDV MTH87 in a human tumor xenograft
The tumor is a highly complex organ that is difficult to accurately model in simple in vitro tissue culture systems. Which factor or combination of factors is missing in the current in vitro tissue culture systems that is critical to selecting for NDV that efficiently replicate, spread and lyse tumor cells is unclear. The human fibrosarcoma cell line HT1080 is a model cell line that is highly sensitive for oncolytic cell killing by NDV MTH68 in vitro. However, in vivo the lysis of HT1080 tumors in mice by MTH68 is incomplete and spread of the viral infection within the tumor is limited to many small areas (data not shown). Therefore, this cell line is well suited to screen for viruses that may have the potential to overcome these in vivo barriers.
We took the simple approach of allowing passage of the virus in vivo to select for variants that had properties that rendered these viruses superior in their in vivo activity relative to the original virus strain. This methodology seeks to take advantage of the natural mutation rate inherent to any replicating organism. For RNA viruses, the mutation rate is estimated to be 0.7 per virus genome per generation.15 Consequently, in a population of 107 virus particles that could be administered in a traditional tumor xenograft model, there should be a calculated population of 0.7 × 107 mutated viral genomes and, assuming that this occurs in a non-prejudiced manner, should create diversity of 7 × 106. We sought to use this natural diversity to allow us to identify highly potent viruses, using the complexity of the tumor to select for highly potent variants in vivo. In our experimental system, tumor xenograft-bearing mice were treated with recombinant NDV harboring the transgene GFP (MTH87).12 Notably, one of the tumors did not grow significantly during the treatment (mouse 3/5, Figure 1a). In contrast, all the vehicle-treated tumor-bearing mice had to be euthanized within 2 weeks after treatment due to tumor burden. It was therefore assumed that a variant or variants in mouse 3/5 replicated and spread efficiently and thus grew better in the tumor than the parental virus that had been used for the treatment.
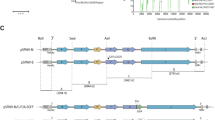
Tumor passaging of oncolytic NDV. (a) Tumor growth curve of HT1080 xenografts in nude mice. Solid line: mean of vehicle-treated animals; dashed line: mean of virus (MTH87)-treated animals; dotted line: individual growth curve mouse 3/5. (b) Scheme of the two rounds of the bioselection procedure. (c) Syncytia formation after infection. Confluent MiaPaCa human tumor cells were infected with mock (top), MTH87 parental virus (middle) or one-time tumor-passaged virus pool (bottom) with an MOI of 0.001. At 24 h after infection, the cells were stained with Giemsa's solution to visualize syncytia.
To prove that replicating virus was still present in the tumor, the tumor was excised from the euthanized animal, dissected into pieces and incubated on a monolayer of HT29 human colon carcinoma cells (Figure 1b). These cells were chosen as indicator cells because they show strong expression of virally expressed GFP after infection with MTH87 before they are killed by the infection. They are also good producer cells to amplify the virus. After incubation with the tumor pieces, GFP-positive cells could be detected (data not shown). This was proof that virus was still present in the tumor and that it was still able to infect the tumor cells and lead to transgene expression. The reisolated viruses were amplified in HT29 producer cells and titrated. Subsequently, the tumor-passaged virus pool was characterized by infection of various tumor cells. When MiaPaCa human pancreatic carcinoma cells that are very sensitive to MTH68, similar to HT1080 cells, were incubated with that virus, a markedly enhanced syncytia formation in the cell monolayer was observed (Figure 1c). This phenotype indicated that bioselection of the virus had selected for at least some phenotypically distinct virus. It also suggested that enhanced syncytia formation correlates with enhanced oncolytic potency in the xenograft.
After the first tumor passage, the virus was amplified in chicken eggs to obtain high-titer stocks for a second round of bioselection. The virus pool was again injected into xenografts of tumor-bearing nude mice. After 32 days, one tumor was again explanted and virus could be reisolated in cell culture on HT29 cells. The supernatant of the HT29 cells was then used to isolate individual viral variants from the pool of bioselected virus MTH87.
Bioselection results in multiple phenotypic distinct virus clones
Individual viruses were isolated by double plaque purification. Out of 48 individual viruses that were isolated and initially analyzed, 8 isolates were selected for expansion and further characterization, as they exhibited individually distinct phenotypes after infection of HT29 cells. Those eight isolates were subjected to a second plaque purification and Figure 2a shows single representative ‘plaques’ after infection of confluent HT29 monolayers with a very low multiplicity of infection (MOI) of 0.0001. It can be observed that isolates 15, 30 and 37 clearly show syncytia formation in contrast to the parental virus MTH87 that does not lead to cell–cell fusion in the HT29 cells. Isolate (cl) 45 demonstrated accelerated spread in the monolayer leading to larger GFP-expressing plaques after 24 h.
Bioselection yields distinct virus clones with different characteristics. (a) HT29 tumor cells were infected with MOI 0.0001 of the parental virus MTH87 and eight selected double plaque-purified isolates of bioselected virus. At 24 h after infection, fluorescence microphotographs were taken of representative individual ‘plaques’ demonstrating the differences in the course of the infection between the individual clones. All clones retained the expression of the transgene GFP. (b) Viral spread: Confluent monolayers of HT29 cells were infected with MOI 0.001 of the indicated viruses. At 0, 24 and 48 h after infection, the cells were brought into suspension by trypsinization and subjected to GFP-FACS analysis. Cells infected by virus and expressing the transgene GFP will score positively. The percentage of GFP-positive cells is plotted against time as an indicator of the spread of the infection. (c) Viral growth rate: Confluent HT29 cells were infected with the indicated viruses. The viral titer was determined during the course of the infection by plaque assay. The histogram shows the maximal titer increase within 24 h, which is the maximal growth rate of the virus in these cells. (d) Cytotoxicity: human tumor cells NCI H460, HT1080 and HT29 were infected with the indicated viruses at an MOI 0.01. The viability of the cells was determined by MTS assay 72 h after infection. The histogram shows the mean value from the experiment in all three cell lines.
Biological characterization of selected clones
To quantify spread of the viral infection, the fraction of GFP-positive cells after the infection was determined by fluorescence-activated cell sorting (FACS) analysis. As seen in Figure 2b, there were different rates of spread for the eight selected isolates. Some isolates exhibited rapid spread in the cell monolayer (15, 45, MTH87), others an intermediate (48, 37, 30, 27, 29) spread and cl 11 a markedly slower spread. Only cl 45 resulted in significant cell death under these experimental conditions, which is reflected by the drop in the number of GFP-positive cells. After 48 h, all clones resulted in more than 70% of the cells being infected and expressing the viral transgene GFP.
The rate of the spread of the infection need not necessarily parallel the rate of virus replication, which measures the increase in virus particles in the cell supernatant. Therefore, also the viral replication was measured for the eight isolates. The maximal increases in infectious virus particles (plaque-forming unit (pfu)) in the supernatant within 24 h after infection of HT29 cells are depicted in Figure 2c. The parental virus MTH87 clearly has the highest growth rate. The isolate cl45, with the fastest microscopically observed spread, has only an intermediate replication rate with regard to virus particle amplification. Isolate cl37, which exhibited the formation of the largest syncytia, had the lowest replication rate.
A traditional indicator of oncolytic potency is in vitro cytotoxicity. To test this property of the individual isolates, confluent monolayers of three different human tumor cell lines (NCI H460 non-small cell lung cancer, HT29 colon carcinoma, HT1080 fibrosarcoma) were infected at a low MOI (0.01). After 72 h, the viability of the cells was determined using an MTS assay. Figure 2d shows the mean viability values of the three cell lines after infection. Several isolates exhibited stronger cytotoxicity than the parental MTH87 virus. From this experiment, it was concluded that the isolates cl 30, cl 37 and cl 45 have the highest in vitro oncolytic potency. Therefore, these three isolates were selected for further analysis and characterization. It should be noted that the three most cytotoxic clones do not have similar properties with respect to fusogenicity, viral spread and growth rate (Table 1).
Increased oncolytic potency in multicellular spheroids and tumors
The faster spread observed with cl 45 may contribute to enhanced oncolytic potency. Also an increased ability to fuse cells, as observed with cl 30 and cl 37, may be an advantage for an oncolytic virus. The parental virus NDV MTH87 is a very cytotoxic virus when applied to cell monolayers.12 Within a tumor tissue, in contrast, the virus is much less destructive and usually not capable of eradicating all tumor cells within a treated xenograft (Beier et al., unpublished data). To test the bioselected virus isolates under more clinically relevant conditions, we used cell spheroids to provide a three-dimensional substrate for the oncolytic virus. HCT116 human colon carcinoma cells can be grown as spheroids with a diameter of up to 400 μm. In monolayer, they are susceptible to MTH87 infection, and though these cells demonstrated strong GFP expression, they did not exhibit significant syncytia formation (data not shown). HCT116 spheroids were infected for several days with either the parental virus MTH87 or the bioselected-derived clones at low MOI (Figure 3). All viruses lead to cell killing. However, the parental virus was less effective than the clones in eliminating the cells in the 3D model. Compared with monolayers, the oncolytic effect is delayed by several days suggesting that the 3D packing of the cells is a barrier to the spread of the virus. Within 2 days after infection, only the rim of the spheroids gets infected and is destroyed. Only after 5–9 days also the center of the spheroids is reached by the virus and eventually killed. The infection with the parental virus MTH87 does not result in a complete cell killing of the spheroids. The result of the cell viability quantification (Figure 3b) is consistent with the microscopic finding confirming that all three tested clones are more oncolytically potent than their parental virus.
Characterization of the bioselected clones in multicellular spheroids. (a) HCT116 cells were grown as spheroids. Spheroids with an average diameter of 1000 μm were infected with MOI=0.01 of the indicated viruses. Individual spheroids were monitored during the course of the infection and microphotographs were taken at the indicated time points with a × 2.5 objective. (b) Quantification of the cytotoxic effect as in (a). Spheroids were dissociated 7 days after the infection and viability was determined using an MTS assay. The histogram shows the viability of the infected cells compared with the mock-treated cells (c) HCT116 cells were grown as large multicellular aggregates with a diameter of ∼3000 μm. Cells were infected with MOI=0.01 of virus and the individual wells were monitored for 8 days. At the indicated times, microphotographs were taken with a × 2.5 objective both in bright field and GFP-fluorescence. The images show the overlay of the two channels to demonstrate the virus spread within the spheroids.
In addition to the spheroid studies, large multicellular aggregates were also generated and used for infection assays. These disc-shaped cell complexes can reach a diameter of 2000 μm. After infection with the GFP viruses for 1 day, a thin layer of cells at the rim of the spheroid is infected and expresses the transgene (Figure 3c). No obvious differences between the viruses can be observed. At 4 days after the infection, virally expressed GFP can clearly be detected also in the center of the aggregates after infection with isolates 37 and 45. The spread of the virus from the rim to the center is delayed after infection with isolate 30 and even more delayed after infection with the parental wild-type virus. It is noteworthy that all viruses were not able to lyse the aggregates, although most of the cells were not viable any more (as assessed by trypan blue staining, data not shown).
Having shown an increased potency of bioselected virus clones in an in vitro 3D tumor cell model, we consequently wanted to test the oncolytic potency of the bioselected viruses also in an in vivo tumor model. Nude mice were transplanted with human HT1080 tumor cells and treated after the tumor area reached 20 mm2 with the isolates cl 30, cl 37, cl 45, as well as the parental strain MTH87. All animals received a dose of 3 × 107 pfu at day 7, 14 and 21. Figure 4 shows the tumor growth curves for the individual animals. All vehicle-treated control animals had to be euthanized by day 14 due to tumor burden (Figure 4, top). MTH87 showed a significant tumor growth inhibition with some animals responding better than others. Treatment with all three individual bioselected isolates resulted in a significantly superior anti-tumor effect than treatment with the parental virus. All animals had a growth inhibition or complete tumor response. Some animals died in the course of the experiment (one animal in the cl 30- and cl 45-treated cohorts, two animals in the cl 37 treatment cohort). These events were observed at time points later than the killing of the control animals due to tumor burden. There was no general loss in body weight in all treatment groups compared with the vehicle-treated group. In contrast to treatment with parental virus, the clones did not only slow down tumor growth but also lead to complete responses, which had not been observed with the wild-type virus. Most responders were seen in the group treated with isolate 45 (4 out of 8), followed by isolate 30 (2 out of 8) and 37 (2 out of 8) and MTH87 (0 out of 8).
In vivo anti-tumor effect. Nude mice were transplanted with HT1080 human xenografts. The tumors were treated intratumoral with a single dose 3 × 107 pfu of either buffer only, the parental virus or the tumor-passaged clones cl 30, cl 37 or cl 45. The diagrams show the growth (tumor area) of the individual tumors in each group (n=8) for the duration of 70 days after treatment. Treatment with the bioselected isolates results in responses: 0 with MTH87, 2 with cl 30, 2 with cl 37 and 4 with cl 45.
It can be concluded that bioselection of MTH87 in human tumor xenografts yielded viruses with enhanced oncolytic potency not only in vitro but also in vivo.
Molecular characterization of the surface proteins of the selected clones
From the observation that some of the bioselected clones differed with regard to syncytia formation, mutations in the genes for the two surface proteins, the fusion protein F and the hemagglutinin neuraminidase protein HN, were most likely to contribute to the differing oncolytic properties. To understand whether particular mutations or variations in those proteins were responsible for the altered phenotypes, F and HN were sequence analyzed and compared with the genes of the parental virus.
It was possible to identify several base substitutions, some of which also result in a change in the amino-acid sequence of the respective protein (summarized in Table 2). From the more fusogenic phenotypic isolates cl 30 and cl 37, we expected to identify differences in the fusion protein. However, changes in the F protein were only identified in cl 30 (Table 2). In contrast, we could not detect any amino acid change in the F proteins of cl 37 and cl 45, although cl 37 exhibited a strong fusogenic phenotype. Therefore, the ability of isolate cl 37 to form more syncytia than parental virus must be independent of the F protein gene sequence.
Amino-acid changes were noted in the HN proteins of all of the studied isolates. One of the changes, T118A, is common to all three isolates, and N119K is shared between cl 37 and cl 45. These relationships suggest that the clones have not emerged completely independently. The common mutation T118A is present in almost all sequenced HN proteins in the Genbank database. D82N is described to be in the F-interacting region in the stem of HN.16 An E495V mutation is described to alter a monoclonal antibody recognition.17 For the other mutations, no function has been reported.
Mutations in the HN protein correlate with the enhanced fusogenicity
From the sequencing results with cl 37, it was clear that differences in the F protein could not be responsible for the differences in the cell–cell fusion behavior between the viruses. For the fusion of cell membranes, it is described that F alone is not sufficient but rather has to cooperate with the HN protein. An interaction between F and HN is very likely necessary.18, 19 Therefore, we wanted to test whether instead of F, the changes in the HN proteins were the cause for enhanced fusogenicity. For that purpose, the genes for the HN proteins of cl 30, cl 37 and cl 45 were cloned as complementary DNA into eukaryotic expression plasmids. Transient transfection of these expression plasmids individually resulted in a very similar staining pattern as transfection with the wt gene (Figure 5a). No obvious differences in expression levels in the cell monolayer, membrane staining or intensity could be observed. Cotransfections of expression plasmids for the fusion and hemagglutinin proteins of paramyxoviruses are described to be sufficient to lead to cell–cell fusion (for example, Sergel et al.19) Cotransfections were performed with combinations of plasmids coding for the wild-type MTH68 F protein and the respective HN mutants of the bioselected virus clone. A clear difference was noted with respect to the cell–cell fusion properties (Figure 5b). Whereas the wild-type HN lead to only small syncytia, all HN proteins derived from the tumor-passaged isolates resulted in significantly larger syncytia. HN from cl 37 demonstrated the greatest increase in fusogenic property, in agreement with the properties already observed with the viral infection in the HT29 cells (Figure 2a). Consequently, individual amino-acid changes in the HN protein are sufficient to account for enhanced fusogenicity of the virus.
Molecular characterization of the HN proteins. (a) U2OS cells were transfected with expression plasmids encoding for the HN protein of the indicated viruses. At 48 h after transfection, cells were stained with an antibody against NDV and a secondary anti-chicken Cy3 antibody as well as Hoechst33258. The red fluorescence shows the expression of the HN protein in the cells, the blue fluorescence shows the cell nuclei of all cells on the coverslip. Pictures were taken with a × 20 objective. (b) U2OS cells were cotransfected with expression plasmids for the indicated proteins. At 48 h after transfection, the cells were fixed and stained with Giemsa's solution to visualize syncytia formation. Pictures were taken with a × 10 objective.
The bioselected viruses have no increased toxicity in the natural host chicken
NDV is a chicken pathogen. The described recombinant virus MTH87 is derived from a mesogenic strain that is classified as an animal pathogen. It is important that viruses designed and selected for oncolytic potency in human tumors do not exhibit an enhanced safety concern for poultry. An increase in virus potency that was coupled with an increase in toxicity to poultry would therefore not be acceptable for a therapeutic oncolytic virus. For that reason, the pathogenicity of the bioselected clones was tested in the natural host chicken. Ten-day-old embryonated chicken eggs were infected at the same MOI for all viral isolates in comparison with the parent virus and the survival of the embryos was monitored twice daily (Figure 6). The highest mortality was seen after infection with MTH87. All bioselected isolates had a similar or better survival rate in the embryos. The calculated mean death time for the isolates is 40–64 h compared with 36 h of the wild-type MTH87. In the case of the isolates cl 30, cl 37 and cl 45, the bioselection in human tumor tissue had therefore not increased the overall toxicity of the virus. Therefore, the toxicity and oncolytic potency of bioselected NDV are not directly linked.
Discussion
The present work demonstrates that in vivo bioselection that seeks to exploit the low mutation rate for paramyxoviruses, which results in microvariants of each strain after only a few genome replications, can be used to isolate highly potent oncolytic NDV viruses. In our study, we demonstrate the power of the methodology to isolate and expand the rare variant that possesses desired, selected-for properties. The rare nature of these isolates support the previous work, that defined the genomic stability of NDV and supports the safety and pharmaceutical utility of the virus.
Although all isolates still expressed the transgene GFP (no colorless syncytia were observed, data not shown), the clones showed marked differences in the spread of the infection and ability to form syncytia, especially when compared with the parental virus. After characterization of a selection of eight bioselected isolates, three isolates showed a marked increase in oncolytic potency when tested on adherent cells. However, no common phenotype could be detected. Whereas isolate cl 37 had a strong fusogenic phenotype, cl 45 was not very fusogenic but spread extremely rapidly within the cell culture. When tumor cells are infected at a very high dose (MOI 1) to assess the oncolytic effect largely independent of virus replication, interestingly cl 45 is almost the least potent of the eight clones (Supplementary Figure S4). This indicates that the main advantage of cl 45 is the fast replication and viral spread, rather than the cytotoxicity. This property may eventually be engineered into one of the other clones to further increase the oncolytic potency.
From this data, it is obvious that various distinct properties of the virus that are the consequence of subtle protein sequence variations contribute similarly to the oncolytic effect. It is well conceivable that it will be possible to engineer even more potent viruses when these independently occurring variations would be combined in a genetically modified virus.
To try to better define the potential clinical activity of these viral variants, we tested their activity on three-dimensional cell spheroids that more closely model the in vivo situation.20, 21, 22 That model has been applied in the present study. The experiments indeed demonstrated that the virus penetration is different for the selected viruses. However, it was observed that although cl 45, which is much less fusogenic than cl 37, had a similar infection pattern and better penetration than wild-type virus. Therefore, the observed effect in the spheroid model cannot exclusively be attributed to the better cell–cell fusion of the isolate. In our experiments, enhanced cytotoxicity in spheroids correlates with enhanced in vivo oncolytic potency supporting the usefulness of the 3D models for studies with oncolytic viruses.
In support of the in vitro experiments, we also describe an increased in vivo anti-tumor effect of the bioselected viruses in a xenograft experiment (Figure 5). The cells that were used for that experiment (HT1080) very easily form syncytia in vitro. Therefore, the more fusogenic phenotype of the isolates may be underestimated and may be more pronounced in less-fusogenic tumor cells, for example, HCT116 or HT29 cells. The clearance of virus from the organism cannot be measured in the in vitro assays. It is therefore possible, that cl 30 and cl 45, which have the highest response rate in vivo (Supplementary Figure S2), have the ability to remain in the organism longer than the parental virus and cl 37. For cl 30 and cl 45, virus could be isolated from the remaining tumors that did not grow 55 days after treatment (data not shown). Exemplified for one tumor, the viral protein expression in the tumor at day 76 was confirmed by immunohistochemistry (Supplementary Figure S3). The molecular basis for that virus persistance remains to be elucidated.
Most responders and the best antitumor effect was observed with cl 45. This isolate is characterized by an extremely fast spread and high toxicity in vitro. It therefore appears that the rate of virus spread from cell to cell is a critical determinant of an effective oncolytic virus. This is in agreement with proposed mathematical models of viral oncolytic therapy,23, 24 which concluded that efficient tumor cell killing is critically dependent on how rapidly the virus lyses infected cells. Our findings now fully support these models experimentally.
The NDV genome encodes six major proteins. Two of them are transmembrane proteins and are expressed on the viral surface: the fusion protein F and the hemagglutinin neuraminidase protein HN. HN is the attachment protein and F is responsible for the fusion of the viral coat with the host cell membrane. When expressed as a transgene in cells, either by transfection or infection, F and HN together lead to syncytia formation in many cells by cell–cell fusion. Some F proteins can promote cell–cell fusion alone. HN alone has no cell fusion activity. If F is expressed together with the homotypic HN-protein, syncytia formation is markedly enhanced.25 The precise mechanism of the fusion-promoting activity of HN is still unknown. A mutation in the NDV F protein in the heptad region 3 (L289A) leads to an increased fusogenicity of the protein.26
To understand the molecular basis for the differences in the fusion behavior of the bioselected viruses, the surface proteins were sequenced. As the fusogenicity of a paramyxovirus is largely determined by the F protein, it was expected that mutations in the F protein were the cause for the observed differences with the bioselected isolates. Strikingly, the most fusogenic isolate cl 37 had no change in the amino-acid sequence of the F protein compared with the parental virus. In contrast, amino-acid changes in the HN protein were discovered. From the transient cotransfection experiments (Figure 5), it is concluded that indeed these mutations in the HN protein are responsible for the enhanced fusogenicity. Because the expression of the HN proteins is not affected, the amino-acid changes must have a mechanistic impact on the cell–cell fusion.
The only common mutation among all three analyzed clones is T118A. Almost all published HN sequences have A at position 118 (BLAST (basic local alignment search tool)-query). Therefore, it is possible that the rare exception 118-T that is present in MTH68, functions to impede cell–cell fusion. A molecular function for aa 259 is not described. When looking at the 3D-structure of HN, the amino acids E259 as well as E495 are on the exposed protein surface.27 However, there is no evidence whether they are involved in the interaction with the F-protein.
Ren et al.28 describe the effect of heptad repeat mutations in F on the HN-dependent fusion and a L289A mutation in F in particular is described to enhance syncytia formation.26 In contrast to F, for the HN protein all amino-acid changes that were identified in this study have not yet been linked with a fusion-promoting function, so this finding may contribute to the elucidation of the role of HN in NDV fusion.
It has been shown that more fusogenic proteins can enhance the oncolytic potency of a virus, for example, the NDV F L289A in VSV,29 suggesting that the ability of an oncolytic virus to form syncytia correlates with the potency. If foreign hyperfusogenic transgenes, such as GALV, are expressed by other recombinant oncolytic viruses it is also possible to enhance the potency of the viruses30, 31, 32 supporting this hypothesis.
These results strongly suggest that cell–cell fusion is of advantage for the oncolytic potency. Our unpublished results from immunuhistochemistry of infected tumors with MTH68 wild-type virus show an incomplete penetration of the infection within the tumor and strongly argue that a major limit of the in vivo efficacy of NDV is the limited intratumoral spread. That issue may be overcome by enhanced fusogenicity of the virus as also supported for vaccinia virus.33 Alternatively, relaxin as a transgene that enhances intratumoral spread is able to increase the oncolytic efficacy of an adenovirus.33, 34
Although the enhanced fusogenicity of the clones can be pinpointed to the mutations in the HN protein, it is still not fully understood what accounts for the enhanced oncolytic effect. Additional mutations in the other genes or the regulatory sequences may contribute. Consequently, future studies will be required to completely sequence these viruses and then systematically separate driver from passenger mutations in the virus genome.
We were able to identify more potent oncolytic viruses by bioselection. To rule out the concern that increased oncolytic potency is also associated with increased toxicity towards normal cells, we infected normal primary fibroblasts with the clones in comparison with the parental virus. There was no significant difference in the cytotoxicity in that experiment (Supplementary Figure S1). The bioselected clones, therefore, maintain the tumor selectivity of the parental virus.
Because we were studying a mesogenic NDV that is classified as an animal pathogen, we were also concerned that the bioselection may have resulted in virus variants that not only replicated better in human tumor but also in their natural host chicken. Although we cannot conclude from the data (Figure 5) that the bioselected clones are more attenuated, we can state that the increased oncolytic potency in human tumor cells does not directly correlate with the toxicity in chicken. This is an important finding because it will not generally limit the bioselection method. In the long term of course, it would be desirable to develop a virus that has no environmental risk any more and is completely attenuated for chicken. Whether the method of bioselection analogously to vaccine research will be useful for that purpose remains to be determined. In general, such a strategy to decrease the chicken toxicity should not automatically decrease concomitantly the oncolytic potency.
We conclude that improved oncolytic viruses compared with the parental virus MTH68 were generated for further development and that the method of bioselection is a useful tool to enhance the properties of an oncolytic paramyxovirus.
Materials and methods
Cells and viruses
HT29 (HTB-38), HT1080 (CCL-121), MiaPaCa (CRL-1420), U2OS (HTB96), NCI-H460 (HTB-177) and HCT116 (CCL-247) cells were obtained from ATCC (LGC Promochem, Wesel, Germany). The virus NDV MTH68 was obtained from Laszlo Csatary, Hungary. MTH87 was described before.12 For in vitro experiments, virus was produced in HT29 cells. Supernatant of cells was centrifuged for 15 min at 3000 r.p.m. and the supernatant was titrated and stored at −80 °C. For in vivo experiments, virus was grown in eggs and allantoic fluid was precleared and concentrated by tangential flow filtration with phosphate-buffered saline (PBS). Concentrated purified virus was titrated and stored in PBS at −80 °C.
Transient transfection of cells was carried out using Lipofectamine 2000 (Invitrogen, Karlsruhe, Germany). In all, 4 × 105 cells were plated in a 6-cm dish and were transfected 1 day later with a total of 1 μg DNA.
For detection of syncytia formation after infection or transfection, cells were fixed with 100% ethanol and stained with 10% Giemsa's solution 24 h after transfection.
For immunofluorescence assay, cells were fixed 2 days after the transfection with 4% formaldehyde. Nuclei of all cells were stained with Hoechst 33258. HN proteins were stained with a chicken polyclonal antiserum against NDV (ab34402, Abcam, Cambridge, UK) and a secondary anti-chicken Cy3 antibody.
Cloning and sequencing
Viral RNA was isolated from purified virus after tangential flow filtration from allantoic fluid using the QIAamp viral RNA kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol. From the resulting RNA, 5 μg were used for each reverse transcription (Superscript kit, Promega, Mannheim, Germany) with gene-specific primers. Subsequently, PCR was carried out with pfu proofreading DNA polymerase (pfu turbo, Stratagene, La Jolla, CA, USA) using primers with restriction enzyme-recognition sequences. The PCR product was cloned into the vector pcDNA2.1 (Invitrogen). The plasmid was used as a template for the sequencing reaction.
Sequencing was carried out with two clones each that originated from two separate first-strand reverse-transcriptase-PCR reactions.
Using the unique restriction sites for SacI and KpnI, the inserts were then subcloned from pcDNA2.1 into the expression vector pCAGGS. Analogously, the gene for the wild-type MTH68 F protein was cloned into the pCAGGS expression plasmid.
Spheroids
The 24-well culture plates were coated with 2% agarose in PBS to prevent adsorbtion of cells. In all, 106 cells were seeded and incubated at 90 r.p.m. on an orbital shaker for the generation of spheroids within 24 h. For generation of larger multicellular aggregates, 106 cells were incubated for 48 h at 70 r.p.m. Cells were infected with 104 pfu. The cells remained on the orbital shaker throughout the experiment.
To determine cell viability with an MTS reagent, the spheroids plus the dissociated cells were transferred into a test tube. The cells were centrifuged for 5 min at 2000 r.p.m. and washed with PBS. The pellet was resuspended in 500 μl trypsin/EDTA to dissociate the spheroid for 5 min at 37 °C. The cells were brought into suspension by pipetting and centrifuged. The pellet was resuspended in 550 μl 37 °C warm medium and split into three times 200 μl in wells of a 96-well plate. A total of 20 μl of MTS reagent (Promega) was added and incubated for 3 h at 37 °C. For safety reasons, the plate was irradiated with UV for 1 h at room temperature to inactivate the virus. The absorbance was read at 490 nm with a reference at 700 nm.
To determine the number of GFP-positive cells within the spheroids, the spheroids were trypsinized as described above. Trypsin was inactivated by addition of fetal calf serum. After centrifugation, the pellet was resuspended in 800 μl 4% formaldehyde and incubated for 15 min at room temperature (fixation and virus inactivation). Cells were centrifuged and resuspended in 800 μl PBS and transferred to 5 ml FACS tubes. In all, 10 000 events were measured in a FACS calibur (Becton-Dickinson, Heidelberg, Germany) for the positivity of GFP fluorescence.
Virus isolation
Tumor-bearing mice were killed by cervical dislocation and the subcutaneous tumor was excised. The tumor was cut in pieces and incubated in culture dishes with confluent HT29 cells in culture medium. When GFP-positive cells were detected, the supernatant was transferred to fresh cells in cell culture flasks. The virus was titrated and 5000 pfu were inoculated into the allantoic cavity of 10-day-old embryonated chicken eggs. After 48 h, the allantoic fluid was harvested and virus was titrated by plaque assay. In parallel, virus was also produced in HT29 cells.
Virus characterization
To measure the spread of the infection 6 × 105 HT29 cells were seeded in 6-well plates. Cells were infected at MOI 0.001 with the respective viruses. After 0, 24 and 48 h cells were trypsinized, fixed with 4% formaldehyde and subjected to FACS analysis to measure the percentage of GFP-positive cells using a FACS calibur (Becton-Dickinson).
The viral growth rate was measured using plaque test titration on HT1080 cells of cell culture supernatant from HT29 cells. HT29 cells were infected at MOI 0.01, MOI 0.001 and MOI 0.0001. An aliquot of cell supernatant was harvested at 0, 24 and 48 h and subjected to titration. The maximal viral growth rate was determined as the maximum increase in viral titer within 24 h.
In vitro cytotoxicity was measured for three different cell lines: human fibrosarcoma HT1080, human colon carcinoma HT29 and human non-small cell lung cancer NCI-H460 tumor cells. Cells were seeded to reach confluency 1 day later in 96-well plates. The cells were infected at an MOI 0.01. After 72 h, cells were incubated with the MTS reagent (Cell Titer 96 AQueous One, Promega) for 2 h at 37 °C. To inactivate virus for safety reasons, the plates were subsequently irradiated with UV for 30 min at room temperature. Absorbance was read at 490 nm, according to the manufacturer's instructions.
Tumor xenograft experiment
Nude mice (Taconic) were inoculated subcutaneously with 3 × 106 cells of HT1080 human fibrosarcoma cells. When tumors reached an average size of 30 mm2, treatment was started. Virus was administered intratumorally with 2.9 × 107 pfu per 100 μl at days 7, 14 and 21. Tumor size and bodyweight of all animals was monitored for 76 days.
Chicken toxicity test
Eleven 10-day-old embryonated chicken eggs (Charles River) were infected with 4000 pfu per 100 μl of cell-derived virus material. The eggs were incubated at 37.5 °C and candled twice daily for 5 days for the survival of the embryos. The number of living embryos was counted at each time point.
References
Fiola C, Peeters B, Fournier P, Arnold A, Bucur M, Schirrmacher V . Tumor selective replication of Newcastle disease virus: association with defects of tumor cells in antiviral defence. Int J Cancer 2006; 119: 328–338.
Roberts MS, Lorence RM, Groene WS, Bamat MK . Naturally oncolytic viruses. Curr Opin Mol Ther 2006; 8: 314–321.
Elankumaran S, Rockemann D, Samal SK . Newcastle disease virus exerts oncolysis by both intrinsic and extrinsic caspase-dependent pathways of cell death. J Virol 2006; 80: 7522–7534.
Alexander DJ . Newcastle disease, other avian paramyxoviruses, and pneumovirus infections. In: Saif YM (ed). Diseases of Poultry. Blackwell Publishing: Ames, Iowa, 2003, pp 63–99.
Sinkovics JG, Horvath JC . Newcastle disease virus (NDV): brief history of its oncolytic strains. J Clin Virol 2000; 16: 1–15.
Hotte SJ, Lorence RM, Hirte HW, Polawski SR, Bamat MK, O’Neil JD et al. An optimized clinical regimen for the oncolytic virus PV701. Clin Cancer Res 2007; 13: 977–985.
Lorence RM, Roberts MS, O’Neil JD, Groene WS, Miller JA, Mueller SN et al. Phase 1 clinical experience using intravenous administration of PV701, an oncolytic Newcastle disease virus. Curr Cancer Drug Targets 2007; 7: 157–167.
Yaacov B, Eliahoo E, Lazar I, Ben-Shlomo M, Greenbaum I, Panet A et al. Selective oncolytic effect of an attenuated Newcastle disease virus (NDV-HUJ) in lung tumors. Cancer Gene Ther 2008; 15: 795–807.
Peeters BP, de Leeuw OS, Koch G, Gielkens AL . Rescue of Newcastle disease virus from cloned cDNA: evidence that cleavability of the fusion protein is a major determinant for virulence. J Virol 1999; 73: 5001–5009.
Romer-Oberdorfer A, Mundt E, Mebatsion T, Buchholz UJ, Mettenleiter TC . Generation of recombinant lentogenic Newcastle disease virus from cDNA. J Gen Virol 1999; 80 (Pt 11): 2987–2995.
Janke M, Peeters B, de Leeuw O, Moorman R, Arnold A, Fournier P et al. Recombinant Newcastle disease virus (NDV) with inserted gene coding for GM-CSF as a new vector for cancer immunogene therapy. Gene Therapy 2007; 14: 1639–1649.
Puhler F, Willuda J, Puhlmann J, Mumberg D, Römer-Oberdörfer A, Beier R . Generation of a recombinant oncolytic Newcastle disease virus and expression of a full IgG antibody from two transgenes. Gene Therapy 2008; 15: 371–383.
Vigil A, Park MS, Martinez O, Chua MA, Xiao S, Cros JF et al. Use of reverse genetics to enhance the oncolytic properties of Newcastle disease virus. Cancer Res 2007; 67: 8285–8292.
Kuhn I, Harden P, Bauzon M, Chartier C, Nye J, Thorne S et al. Directed evolution generates a novel oncolytic virus for the treatment of colon cancer. PLoS ONE 2008; 3: e2409.
Drake JW, Holland JJ . Mutation rates among RNA viruses. Proc Natl Acad Sci USA 1999; 96: 13910–13913.
Deng R, Wang Z, Mahon PJ, Marinello M, Mirza A, Iorio RM . Mutations in the Newcastle disease virus hemagglutinin-neuraminidase protein that interfere with its ability to interact with the homologous F protein in the promotion of fusion. Virology 1999; 253: 43–54.
Gotoh B, Sakaguchi T, Nishikawa K, Inocencio NM, Hamaguchi M, Toyoda T et al. Structural features unique to each of the three antigenic sites on the hemagglutinin-neuraminidase protein of Newcastle disease virus. Virology 1988; 163: 174–182.
Lamb RA . Paramyxovirus fusion: a hypothesis for changes. Virology 1993; 197: 1–11.
Sergel T, McGinnes LW, Peeples ME, Morrison TG . The attachment function of the Newcastle disease virus hemagglutinin-neuraminidase protein can be separated from fusion promotion by mutation. Virology 1993; 193: 717–726.
Lamfers ML, Hemminki A . Multicellular tumor spheroids in gene therapy and oncolytic virus therapy. Curr Opin Mol Ther 2004; 6: 403–411.
de Graaf M, Pinedo HM, Oosterhoff D, van der Meulen-Muileman IH, Gerritsen WR, Haisma HJ et al. Pronounced antitumor efficacy by extracellular activation of a doxorubicin-glucuronide prodrug after adenoviral vector-mediated expression of a human antibody-enzyme fusion protein. Hum Gene Ther 2004; 15: 229–238.
Lam JT, Hemminki A, Kanerva A, Lee KB, Blackwell JL, Desmond R et al. A three-dimensional assay for measurement of viral-induced oncolysis. Cancer Gene Ther 2007; 14: 421–430.
Wodarz D, Komarova N . Towards predictive computational models of oncolytic virus therapy: basis for experimental validation and model selection. PLoS ONE 2009; 4: e4271.
Wein LM, Wu JT, Kirn DH . Validation and analysis of a mathematical model of a replication-competent oncolytic virus for cancer treatment: implications for virus design and delivery. Cancer Res 2003; 63: 1317–1324.
Horvath CM, Paterson RG, Shaughnessy MA, Wood R, Lamb RA . Biological activity of paramyxovirus fusion proteins: factors influencing formation of syncytia. J Virol 1992; 66: 4564–4569.
Sergel TA, McGinnes LW, Morrison TG . A single amino acid change in the Newcastle disease virus fusion protein alters the requirement for HN protein in fusion. J Virol 2000; 74: 5101–5107.
Crennell S, Takimoto T, Portner A, Taylor G . Crystal structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase. Nat Struct Biol 2000; 7: 1068–1074.
Ren G, Wang Z, Wang G, Song Y, Yao P, Xu H et al. Effects of heptad repeat regions of F protein on the specific membrane fusion in paramyxoviruses. Intervirology 2006; 49: 299–306.
Ebert O, Shinozaki K, Kournioti C, Park MS, García-Sastre A, Woo SL . Syncytia induction enhances the oncolytic potential of vesicular stomatitis virus in virotherapy for cancer. Cancer Res 2004; 64: 3265–3270.
Fu X, Tao L, Jin A, Vile R, Brenner MK, Zhang X . Expression of a fusogenic membrane glycoprotein by an oncolytic herpes simplex virus potentiates the viral antitumor effect. Mol Ther 2003; 7: 748–754.
Johnson KJ, Peng KW, Allen C, Russell SJ, Galanis E . Targeting the cytotoxicity of fusogenic membrane glycoproteins in gliomas through protease-substrate interaction. Gene Therapy 2003; 10: 725–732.
Simpson GR, Han Z, Liu B, Wang Y, Campbell G, Coffin RS . Combination of a fusogenic glycoprotein, prodrug activation, and oncolytic herpes simplex virus for enhanced local tumor control. Cancer Res 2006; 66: 4835–4842.
Kim JH, Lee YS, Kim H, Huang JH, Yoon AR, Yun CO . Relaxin expression from tumor-targeting adenoviruses and its intratumoral spread, apoptosis induction, and efficacy. J Natl Cancer Inst 2006; 98: 1482–1493.
Ganesh S, Gonzalez Edick M, Idamakanti N, Abramova M, Vanroey M, Robinson M et al. Relaxin-expressing, fiber chimeric oncolytic adenovirus prolongs survival of tumor-bearing mice. Cancer Res 2007; 67: 4399–4407.
Acknowledgements
We thank Katja Köckritz for excellent technical assistance and Florian Pühler for critical reading of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Rudolf Beier, Dominik Mumberg and Terry Hermiston are employed by Bayer Healthcare Pharmaceuticals.
Additional information
Supplementary Information accompanies the paper on Gene Therapy website
Rights and permissions
About this article
Cite this article
Beier, R., Hermiston, T. & Mumberg, D. Isolation of more potent oncolytic paramyxovirus by bioselection. Gene Ther 20, 102–111 (2013). https://doi.org/10.1038/gt.2012.13
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gt.2012.13