Abstract
Purpose
Constitutional epimutations are an alternative to genetic mutations in the etiology of genetic diseases. Some of these epimutations, termed secondary, correspond to the epigenetic effects of cis-acting genetic defects transmitted to the offspring following a Mendelian inheritance pattern. In Lynch syndrome, a few families with such apparently heritable MLH1 epimutations have been reported so far.
Methods
We designed a long-range polymerase chain reaction next-generation sequencing strategy to screen MLH1 entire gene and applied it to 4 French families with heritable epimutations and 10 additional patients with no proven transmission of their epimutations.
Results
This strategy successfully detected the insertion of an Alu element in MLH1 coding sequence in one family. Two previously unreported MLH1 variants were also identified in other epimutation carriers: a nucleotide substitution within intron 1 and a single-nucleotide deletion in the 5′-UTR. Detection of a partial MLH1 duplication in another family required multiplex ligation-dependent probe amplification technology. We demonstrated the segregation of these variants with MLH1 methylation and studied the functional consequences of these defects on transcription.
Conclusion
This is the largest cohort of patients with MLH1 secondary epimutations associated with a broad spectrum of genetic defects. This study provides further insight into the complexity of molecular mechanisms leading to secondary epimutations.
Similar content being viewed by others
Introduction
A constitutional epimutation is an epigenetic defect that causes disruption of gene expression and is present in the DNA of normal tissues. It is restricted to one allele of the gene and can exhibit mosaicism. Epimutations have been identified as an alternative etiological mechanism to genetic mutations in a few human diseases, such as Lynch syndrome,1,2,3 α-thalassemia,4 and breast and ovarian cancer predisposition.5
Epimutations are considered to be nonheritable as epigenetic marks are erased in gamete precursors and in the early embryo.6 Nevertheless, families with several epimutation-carriers have been described.1,7,8,9,10 Two types of constitutional epimutations have been defined:2 primary epimutations corresponding to pure epigenetic events, labile in the germ line and thus reversible between generations (although non-Mendelian inheritance might be occasionally observed11), and secondary epimutations corresponding to the secondary epigenetic effects of cis-acting genetic alterations transmitted following a Mendelian inheritance pattern, with re-establishment of the epigenetic change in the offspring.
Constitutional epimutations of MLH1 or MSH2 genes have been identified as a rare etiology of Lynch syndrome.2,12,13 Lynch syndrome is an autosomal dominant genetic condition predisposing to colorectal, endometrial, and other extracolonic cancers.14 It is caused by germ-line mutations in one of the DNA mismatch repair (MMR) genes, MLH1, MSH2, MSH6, or PMS2. Tumors exhibit MMR deficiency, which is the consequence of somatic inactivation of the second allele of the affected gene and leads to instability of microsatellite sequences in the tumor genome.
Epimutations of MLH1 and MSH2 correspond to methylation of CpG sites located in the CpG islands within promoter regions, leading to transcriptional silencing of the affected allele. MSH2 epimutations are typical secondary epimutations, being the consequence of germ-line deletions of the 3′ end of the EPCAM gene, which is located 17 kb upstream of MSH2. Loss of EPCAM polyadenylation signal leads to production of EPCAM-MSH2 fusion transcripts and epigenetic silencing of MSH2, especially in epithelial cells where EPCAM promoter activity is high.15 Thus, great variations of methylation levels are commonly observed between peripheral blood lymphocytes (PBLs) (very low methylation level) and epithelial cells such as colonic cells (high methylation level).1 In contrast, MLH1 epimutations can be primary or secondary. MLH1 hypermethylation is observed in various cell types throughout the body, although with some degree of somatic mosaicism. In contrast to MSH2, molecular mechanisms underlying the establishment of hypermethylation of MLH1 promoter CpG remain mostly unexplained.
The first genetic defect associated with MLH1 promoter methylation was reported in 2009,16 and so far only a few families exhibiting a secondary epimutation of MLH1 have been described.7,9,17,18 In an attempt to further explore molecular mechanisms leading to constitutional epimutations, we designed a long-range polymerase chain reaction (PCR) next-generation sequencing (NGS) strategy to screen MLH1 entire gene and surrounding sequences and applied it to four French families with heritable epimutations and 10 additional patients with no proven transmission of their epimutations.
Materials and methods
Patients
Thirty-one patients were included in this study: 21 patients from four French families with heritable epimutations (11 epimutation carriers and 10 noncarriers) and 10 additional patients with no proven transmission of their epimutations (Table 1). All patients signed an informed-consent form for genetic analyses.
Long-range PCR and NGS
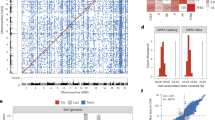
Primers were designed to amplify a 75,365-bp genomic region from chr3:37,025,250 to chr3:37,100,615 (GRCh37/hg19), encompassing EPM2AIP1 and MLH1 genes and the last four exons of LRRFIP2. This genomic region was PCR-amplified, using 13 overlapping long-range PCR reactions (Figure 1a). Libraries were prepared according to the protocol detailed in the Supplementary Methods online and sequenced on a GS Junior system (Roche, Mannheim, Germany).
(a) Long-range PCR amplicons for next-generation sequencing of the region of interest on chromosome 3 (EPM2AIP1: NM_014805.3; MLH1: NM_000249.3; LRRFIP2: NM_006309.3) (Integrative Genome Viewer, Broad Institute). (b) Next-generation sequencing data analysis pipeline for structural variant detection: after mapping of reads to chromosome 3, both unmapped and chimeric reads were extracted and submitted to de novo assembly. Contigs were then blasted against hg19 genome reference, enabling detection of insertion or deletion.
The data analysis (single-nucleotide variant, small indel, and structural variant detection) is described in the Supplementary Methods online and in Figure 1b.
Screening for large genomic rearrangements
The multiplex ligation-dependent probe amplification (MLPA) P003 kit from MRC-Holland (Amsterdam, The Netherlands) was used following the manufacturer’s instructions to screen DNA samples for large genomic rearrangements in MLH1. Characterization of the one identified in family 2 is described in the Supplementary Methods online.
DNA methylation analysis by pyrosequencing
The pyrosequencing assays targeted CpG sites within the MLH1 C- and D-regions reported by Deng et al.19 and within MLH1 intron 1. Methylation of CpG within the Deng-C and -D regions has been strongly correlated with transcriptional silencing.19 For the Deng-C region located in MLH1 promoter, we used the PyroMark MLH1 kit (Qiagen, Courtaboeuf, France) (Supplementary Figure S1 online). We designed new pyrosequencing assays for the Deng-D region located in MLH1 5′-UTR, and for 5 CpG located within intron 1 (inside the CpG island and outside of intron 1 Alu sequences) (Supplementary Figure S1,Table S1 online) using the Biotage PSQ Assay Design software v1.0.6 (Qiagen).
Primer pairs were also designed for allele-specific amplification of bisulfite-treated DNA when possible (i.e., for patients carrying a mutation or heterozygous for the c.-93G>A common single-nucleotide polymorphism) (primer sequences available on request). Controls corresponding to epimutation-negative relatives homozygous for the wild-type allele or to individuals homozygous at the c.-93 nucleotide position (G/G or A/A) were included. Nested PCR was subsequently performed with the pyrosequencing amplification primers using the allele-specific PCR products as templates.
Allele-specific pyrosequencing was also performed for the analysis of intron 1 methylation in family 3, using sequencing primers specific of the c.116+106G or the c.116+106A allele (primer sequences available on request).
PCR and pyrosequencing protocols, as well as molecular methods commonly used in genetics labs, such as Sanger sequencing, fragment length analysis, complementary DNA preparation, and amplification and haplotype analysis, are presented in the Supplementary Methods online.
Results
We developed a long-range PCR NGS strategy to extensively screen MLH1 gene for all types of sequence and structural variations (Figure 1). Using this strategy, we sequenced DNA from patients from four French families with heritable MLH1 epimutations and from patients with no proven transmission of their MLH1 epimutations. The aim was to identify genetic defects associated with transmission of MLH1 epimutations.
Family 1
We previously reported family 1 as the first French family with a secondary epimutation7 (Figure 2a). The putative underlying genetic defect in cis remained unknown. Our long-range PCR NGS and de novo assembly strategy enabled identification of the insertion of an AluYc sequence, 6 bp upstream from the 3′ end of MLH1 exon 1 (Figure 2b): NC_000003.11:g.37035148_37035149ins[NC_000006.11:g.7717384_7717467;7717491_ 7717705;AAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA;NC_000003.11:g.37035135_37035148]. The insertion was 345 nucleotides long. Its sequence was validated by Sanger sequencing. Fragment length analysis was also used to determine the number of adenine repeats at the end of the inserted sequence. It corresponded to a typical Alu element20 with the A5TACA5 sequence separating the left and the right monomers, the poly(A) tail at the 3′ end as a remnant of the RNA intermediate and the target site duplication of flanking region used for insertion in the host genome. The allele with Alu insertion segregated with hypermethylation in the family.
(a) Pedigree of family 1 showing autosomal dominant inheritance of constitutional MLH1 epimutation. The two alleles of individuals who were tested for MLH1 methylation are depicted, with wild-type (WT) allele(s) in gray and methylated allele in purple. The proband is indicated by an arrow; roman numbers are for generations and Arabic numbers are for individuals; squares are males; circles are females; black shading indicates individuals affected by cancer as listed below, with the age of onset also indicated (CRC, colorectal cancer); crossed squares or circles are for deceased individuals. (b) Sequence of exon 1 with insertion of an AluYc sequence. The sequence is in capital letters, with exon 1 in gray and the inserted sequence in bold. Alignment to AluYc sequence using RepeatMasker (http://www.repeatmasker.org/) is depicted in lowercase letters below the sequence (“i” indicates a transition and “−” a deletion). The c.1 nucleotide is the A of the translational initiation codon (GenBank NM_000249.3). The target site duplication (TSD) is indicated in red arrows. The new splicing donor site, as identified by RNA analysis, is indicated by a star. (c) Sequencing of complementary DNA generated from RNA extracted from patient IV-1 lymphoblastoid cell line. Electropherogram shows splicing from a donor site located within the Alu sequence (the last five nucleotides of the forward primer used for polymerase chain reaction (PCR) amplification are complementary to the first five nucleotides of the inserted sequence; the reverse primer is located in exon 4) with expression of a major transcript, and of a minor transcript lacking the first five nucleotides of exon 2. (d) Methylation analysis of C- and D-regions for both alleles and after allele-specific amplification of the WT and the mutant alleles. Positive results are the mean of three independent experiments (three separate bisulfite conversions and PCR amplifications) and standard deviations are indicated. Amplification of the mutant allele is restricted to patients exhibiting the insertion (II-2, III-2, and IV-1), as shown by gel electrophoresis (NTC, no template control). Numbers refer to the ones indicated on the pedigree of family 1.
Analysis of RNA extracted from patients II-2, III-2, and IV-1 lymphoblastoid cell lines showed expression of two different transcripts generated by splicing from a donor splice site located in the Alu sequence, 68 bp from its 5′ end: a major transcript that corresponded to the use of the natural acceptor splice site of intron 1 (r.112_116delinsGCCGGGCGCGGTGGCTCACGCCTGTAATCCCAGCACTTTGGGAGGCCGAGGCGGGCGGATCACGAG), and a minor transcript that corresponded to the use of a cryptic acceptor site located in exon 2, 5 bp after the natural one (r.112_121delinsGCCGGGCGCGGTGGCTCACGCCTGTAATCCCAGCACTTTGGGAGGCCGAGGCGGGCGGATCACGAG) (Figure 2c). Those two transcripts both encoded the same truncated protein, p.(Asn38Alafs*9).
Methylation analyses of MLH1 C- and D-regions after allele-specific amplification determined that hypermethylation was monoallelic and restricted to the allele containing the insertion (Figure 2d). Methylation extended on the inserted sequence and on intron 1 on this mutant allele (data not shown).
Family 2
The proband presented with colorectal cancer (CRC) exhibiting microsatellite instability and apparent isolated loss of PMS2 protein expression, at the age of 52. There was no history of cancer in his first-degree relatives (Figure 3a). Genetic testing of PMS2 gene was performed, but no mutation was identified. MLH1 promoter hypermethylation was detected in tumor DNA and in DNA extracted from PBLs, leading to the diagnosis of constitutional epimutation. Epigenetic testing was offered to other family members and the epimutation was identified in six asymptomatic relatives (i.e., the mother’s proband, one brother, one nephew, and three cousins), thus validating dominant inheritance of the epimutation. A cousin underwent prophylactic oophorectomy and an ovarian carcinoma exhibiting loss of MLH1 and PMS2 protein expression was diagnosed.
(a) Pedigree of family 2 showing autosomal dominant inheritance of constitutional MLH1 epimutation. The two alleles of individuals who were tested for MLH1 methylation are depicted, with wild-type allele(s) in gray and methylated allele in purple. The proband is indicated by an arrow. CRC, colorectal cancer; OvCa, ovarian carcinoma. (b) Identification of a duplication encompassing MLH1 exons 1 to 6 as assessed by multiplex ligation-dependent probe amplification (left panel) and extending to EPM2AIP1 in 5′ as assessed by Agilent CGH Microarray (right panel). (c) Characterization of the breakpoints and schematic representation of the duplication encompassing EPM2AIP1 and MLH1 exons 1 to 6. Genomic locations of the breakpoints are indicated (GRCh37/hg19). The same six-nucleotide sequence (TACAGG) is present at both ends of the duplicated segment. (d) Sequence comparison and alignment between the region containing the breakpoint (middle line, red capital letters) and the sequences involved in the recombination process: MLH1 intron 6 (GRCh37/hg19 chr3:37,051,302-37,051,599) (green capital letters) and the sequence upstream of EPM2AIP1 (GRCh37/hg19 chr3:37,021,762-37,022,052) (blue capital letters). Sequence similarities are indicated by vertical bars. Boxes show perfect homology between the sequences upstream and downstream of the breakpoint. The six nucleotides (TACAGG) found at both ends of the duplicated segment are in bold. The green and the blue sequences have an overall homology of 82% (244/298). Alignment to AluSq2 of nucleotides 37,051,302 to 37,051,599 is depicted in lowercase letters above the green sequence; alignment to AluSx3 of nucleotides 37,021,762 to 37,022,052 is depicted in lowercase letters below the blue sequence (“i” indicates a transition, “v” a transversion, and “−” a deletion) (http://www.repeatmasker.org/). (e) Complementary DNA sequencing and schematic representation of MLH1 transcript expressed from the allele with duplication. Primers for RT-PCR amplification are indicated by arrows (forward primer located in exon 5 and reverse primer located in exon 4). RNA was extracted from lymphoblastoid cell lines from patient III-13 (no difference was observed between puromycin-treated and nontreated cell lines). (f) Methylation analysis of MLH1 C-region, D-region, and IVS1 (allele-specific amplification was not possible due to the size of the duplication). Positive results are the mean of three independent experiments (three separate bisulfite conversions and PCR amplifications) and standard deviations are indicated. Numbers refer to the ones indicated on the pedigree of family 2.
A large duplication encompassing MLH1 exons 1 to 6 and extending to EPM2AIP1 in 5′ was found using MLPA and comparative genomic hybridization (CGH)-array (Figure 3b). Breakpoints were further characterized by quantitative PCR and Sanger sequencing (Figure 3c). This tandem duplication comprised 29.54 kb: NC_000003.11:g.37021898_37051437dup (GRCh37/hg19). Both ends of the duplicated segment were located in Alu elements: an AluSx sequence upstream of EPM2AIP1 gene and an AluSq sequence in MLH1 intron 6, in the same orientation (Figure 3d). A homologous sequence of six nucleotides (TACAGG) was found at both ends. Due to its size, this duplication could not be identified by our long-range PCR NGS strategy.
Analysis of RNA extracted from the proband’s lymphoblastoid cell lines showed that transcription could start from the duplicated segment and read through into exon 2 of the downstream intact copy of the gene, producing an aberrant transcript (Figure 3e). This transcript contained an in-frame insertion of 429 nucleotides after position r.545, which is predicted to result in a protein with an in-frame insertion of 143 amino acids between glycine at position 181 and arginine at position 182 (p.(Gly181_Arg182ins143)). However, duplication encompassed MLH1 promoter and transcription could also start from the second promoter on the rearranged allele, leading to expression of an intact wild-type transcript. Microsatellite instability in the tumors of the proband and of patient III-1 was indicative of MMR deficiency and thus argued against expression of the wild-type transcript only from the variant allele.
The duplication segregated with hypermethylation of MLH1 C and D-regions and intron 1 in the family (Figure 3f). Allele-specific amplification of the MLH1 5′ end on bisulfite-treated DNA was not possible due to the size of the duplication. Nor was it possible to determine which of the two copies of the potentially methylated C- and D-regions on the duplicated allele was actually methylated (they may also both be partially methylated). However it can be assumed that hypermethylation contributes to decreased expression of the intact transcript, leading to the tumor phenotype.
Family 3
MLH1 promoter hypermethylation was first identified in genomic DNA of a female patient, diagnosed with CRC at the age of 58 (patient III-3), and her niece, diagnosed with metastatic CRC at the age of 35 (patient IV-1) (Figure 4a). Patient III-1, as the brother of patient III-3 and the father of patient IV-1, was an obligate carrier and was referred to genetic counseling. His personal history was urothelial carcinoma at the age of 61 and colonic adenomas since the age of 55. Interestingly, the methylation level in DNA extracted from his PBLs was very low (Figure 4b). The epimutation was also detected in her asymptomatic daughter (patient IV-2).
(a) Pedigree of family 3 showing autosomal dominant inheritance of constitutional MLH1 epimutation. The two alleles of individuals who were tested for MLH1 methylation are depicted, with wild-type allele(s) in gray and methylated allele in purple. The two probands are indicated by an arrow. CRC, colorectal cancer. (b) Methylation analysis of MLH1 C-region for family 3 patient III-1: pyrogram showing low methylation level. (c) Haplotype analysis using microsatellites in family 3 and for patient P4 and her daughter. The different alleles of each microsatellite were numbered according to their increasing numbers of repeats. The haplotype associated with the epimutation (Me, methylation) is highlighted in yellow for family 3 and in orange for patient P4. Other haplotypes shared by two first-degree relatives are indicated by different shades of gray. (d) and (e) Methylation analysis of C- and D-regions for both alleles and after allele-specific amplification of the c.116+106G and the c.116+106A alleles (as shown by gel electrophoresis) (NTC, no template control). Positive results are the mean of at least three independent experiments (separate bisulfite conversions and polymerase chain reaction (PCR) amplifications) and standard deviations are indicated. The high variability between replicates and the nonquantitative results observed for the c.116+106A allele may be explained by the requirement for a degenerated reverse primer (primer overlapping 2CpG). (d) Family 3 (numbers refer to the ones indicated on family 3 pedigree). (e) Patient P4. (f) Patient P4: pyrograms showing methylation level of the C-region for both alleles and after allele-specific amplification of the c.-93G allele or the c.-93A allele. The c.-93A and the c.116+106A variants are located on the same allele (same reads in next-generation sequencing results). (g) and (h) Methylation analysis of intron 1 for both alleles and after allele-specific pyrosequencing of the c.116+106G and the c.116+106A alleles. Positive results are the mean of at least three independent experiments (separate bisulfite conversions and PCR amplifications) and standard deviations are indicated. (g) Family 3 (numbers refer to the ones indicated on family 3 pedigree). (h) Patient P4.
In this family, no structural rearrangement of MLH1 gene could be found with our NGS strategy, nor with MLPA. Nevertheless, analysis of sequencing data identified a substitution within MLH1 intron 1 in patients III-1, III-3, and IV-2: c.116+106G>A (NM_000249.3). This variant segregated with hypermethylation in the family. Interestingly, this variant has not been recorded in public databases and it has never been identified in Lynch syndrome routine diagnosis in our lab (380 patients tested by NGS and also tested negative for promoter hypermethylation). In contrast, 1 of the 10 patients with no proven transmission of their epimutations who were screened in this study was also a carrier of the variant (P4). Epigenetic testing was offered to patient P4’s relatives. Her daughter, asymptomatic at the age of 31, and two cousins diagnosed with colorectal cancer, one at the age of 56 (with isolated loss of MSH6 protein expression) and one above 70, were tested, but none was a carrier of the epimutation, nor of the variant c.116+106G>A. Haplotype analysis was performed in the two families. No shared haplotype associated with the c.116+106A allele was identified (Figure 4c).
Analysis of RNA extracted from patients III-1, IV-2, and P4 lymphoblastoid cell lines showed no effect of the variant c.116+106G>A on splicing. As patients III-1 and P4 were heterozygous carriers of MLH1 common single-nucleotide polymorphism c.655A>G, it was possible to check that transcripts were expressed from both alleles of the gene (data not shown).
Methylation analyses of C- and D-regions after allele-specific amplification demonstrated that hypermethylation was monoallelic and restricted to the c.116+106A allele in family 3 and patient P4 (Figure 4d). Patient P4 was a heterozygous carrier of the common single-nucleotide polymorphism c.-93G>A, allowing validation of monoallelic C-region hypermethylation associated with the c.116+106A allele (Figure 4f). Methylation also extended on intron 1 only on this allele, as assessed by allele-specific pyrosequencing (Figure 4g).
Family 4
Family 4 was a large family fulfilling Amsterdam I criteria, exhibiting low-level MLH1 hypermethylation and carrying the synonymous c.27G>A variant previously described in a Caucasian family.21 NGS was performed to search for an additional genetic defect in cis, including complex rearrangement, but none was identified.
Nine patients from three generations were tested in this family (Supplementary Figure S2a online). Five carried the c.27G>A variant and low-level methylation (mean methylation level ≤10% for the C-region and <6.5% for the D-region) (Supplementary Figure S2b online). The other four relatives did not harbor the Me27A allele. The methylated allele segregated with disease. Only the youngest patient with the Me27A allele (aged 26) did not develop colon cancer or adenoma (patient III-4). Interestingly, despite the low-level of methylation, CRC occurred at a relatively young age in this family (below 50) (patients I-4, II-4, and I-6, who is an obligate carrier). Moreover patient I-4 developed 3 CRC despite the quasi-undetectable methylation of DNA extracted from her PBLs, as assessed by pyrosequencing.
In this family, data confirmed transmission of low-level methylation segregating with the c.27G>A variant and allele-specific methylation on the A allele (Supplementary Figure S2c online), as previously reported.21
Another patient of the cohort, a female diagnosed with CRC and endometrial cancer at the age of 49 and 52 (patient P5), also carried the c.27G>A variant. Mean methylation level was 13.5% for the C-region and 8% for the D-region (data not shown). She had a family history of cancer as her brother died from CRC at the age of 30. Only her asymptomatic son could be tested. He was not a carrier of the methylated allele, nor of the c.27G>A variant. The c.27G>A variant was on a different haplotype than that of family 4, as it was associated with the -93A allele for patient P5 whereas it was associated with the -93G allele in family 4.
Other patients
Apart from these four families, 10 patients with no proven transmission of their epimutations were also screened in this study (Table 1). One patient (P4) was a carrier of the c.116+106G>A variant identified in family 3, and another one (P5) was a carrier of the c.27G>A variant identified in family 4.
Sequencing of MLH1 entire gene enabled identification of another variant located in MLH1 5′-UTR in a female diagnosed with CRC at the age of 36 (patient P3): c.-167delA. This variant is not in germ-line databases and it has never been identified in routine diagnosis in our lab. We demonstrated that hypermethylation of the C-region was associated with the -167delA allele, as assessed by allele-specific methylation analysis (Supplementary Figure S3 online).
Discussion
Only a few families with a heritable MLH1 epimutation (i.e., secondary constitutional epimutation) and the underlying genetic defect have been reported so far. In a Finnish family, constitutional MLH1 hypermethylation was associated with a 6.4-kb deletion encompassing exons 1 and 2, thus providing the first evidence that genetic disruption of MLH1 gene can induce epigenetic modifications.16 In another family from Germany, hypermethylation was linked to a large duplication including the entire MLH1 gene and four additional flanking genes.8,17 In five Caucasian families, MLH1 methylation segregated with a large common haplotype harboring two single-nucleotide variants: c.-27C>A and c.85G>T, the c.-27C>A variant associated with reduced transcriptional activity in somatic cells being the most likely candidate.9,21,22 An Italian family with promoter hypermethylation and a 997-bp deletion in cis (c.-168_116+713del) has also been reported.18 The only common feature of these various genetic defects is the loss of integrity of the MLH1 5′-UTR region.
We previously reported a French family with constitutional MLH1 epimutation, but failed to identify the cis-acting underlying genetic defect using conventional molecular methods.7 In the present study, we took advantage of the relatively long reads generated by the GS Junior sequencing platform (average read length: 400–500 bp) and performed de novo assembly on unmapped and chimeric reads to detect complex structural variants. This allowed identification of an AluYc sequence in MLH1 exon 1 in this family. This insertion causes disruption of MLH1 gene and provides a new splicing donor site, promoting alternative splicing. Short interspersed nuclear elements of the Alu class, with more than 1 million copies, are the most abundant repetitive sequences in the human genome, and are mostly present in noncoding regions. They have spread over the whole genome by retrotransposition during evolution and represent a great source of genetic variability. They can cause human genetic diseases through recombination between them or through de novo insertion.20 De novo insertions may occur in the coding sequence of a gene, disrupting the open reading frame.23 A similar mutational mechanism as the one we describe here has been reported in Alström syndrome, with the exonic insertion of an AluYa5 element in ALMS1 gene and the same long uninterrupted stretch of adenosines, which suggests that the Alu sequence was inserted by a recent retrotransposition event.24 Hotspots for the insertion of Alu elements have been identified in the coding sequence of a few cancer genes such as PTEN 25 and NF1.26 In MMR genes, Alu-mediated recombination events represent a well-known mechanism of exonic rearrangements, especially in MSH2 gene, which exhibits the highest density of Alu repeats.17,27,28,29,30 The only report of an Alu insertion in the coding sequence of a MMR gene also concerned MSH2.31 In that report, the 184-bp sequence only represented the 3′ part of an AluJ element, pointing to Alu-mediated recombination as the cause of the insertion and arguing against retrotransposition. We describe here for the first time the insertion into the coding sequence of a MMR gene of a full-length Alu element, probably mediated by retrotransposition, this hypothesis being consistent with activity of AluY, the youngest Alu lineage, in the human genome.32Alu elements are frequently methylated in normal cells33,34 and can act in cancers as methylation centers from which DNA methylation spreads into gene promoters.35 This can explain the establishment of promoter hypermethylation in family 1.
The large duplication identified in family 2 also involves Alu elements: an AluSx located upstream from EPM2AIP1 gene and an AluSq located in MLH1 intron 6, both in the same orientation. This is indicative of a duplication resulting from Alu-mediated homologous recombination. Transcription can start from the duplicated segment and read through into exon 2 of the downstream intact copy of the gene, producing an aberrant transcript. A very similar copy-number variant–based epigenetic scenario has been described previously for the tumor suppressor gene PTPRJ, with a duplication affecting its 5′ end, expression of a read-through transcript, and aberrant promoter methylation.36
In this study we also report a variant located in MLH1 intron 1 (c.116+106G>A). This variant segregates with hypermethylation in family 3, is present in another unrelated epimutation carrier, and is absent from databases, pointing to a potential role in MLH1 promoter methylation.
In family 4, the only relevant genetic variant that we could identify segregating with promoter methylation was c.27G>A. Transmission of low-level methylation in association with this variant has been reported previously, but another genetic basis for cancer predisposition in that family could not be excluded as the Me27A allele did not segregate with disease.21 This variant was also found in patient P5, on a haplotype different from that of family 4.
We also identified a new MLH1 5′-UTR variant on the methylated allele of another patient (c.-167delA). There is no evidence that this epimutation is secondary because no patient’s relative could be tested. But this variant has not been reported previously in germ-line databases and it has never been identified in our lab.
The c.-27C>A and c.85G>T haplotype, previously reported as associated with constitutional MLH1 epimutation in five Caucasian families,9,21,22 was not found in the patients of this study.
With this study, we increase the number of families reported with a secondary epimutation and extend the spectrum of underlying genetic defects to a partial duplication of MLH1 and an Alu insertion, two mutational mechanisms that have not been described previously, and maybe also to an intronic variant and a new single-base deletion in 5′-UTR. This emphasizes the diversity of genetic events causing methylation of MLH1 promoter (including single-nucleotide variants, copy-number variants, structural variants, and, potentially, small indels).
Our data confirm that genetic disruption at the start of MLH1 gene can give rise to an altered epigenetic state, transmitted to the offspring as a somatic mosaic. Epigenetic reprogramming during gametogenesis and early preimplantation embryogenesis consists of two distinct rounds of genome-wide DNA demethylation followed by de novo methylation.6 Erasure of MLH1 methylation in the gametes has been demonstrated in spermatozoa from a patient with a secondary epimutation.9 Methylation marks are then re-established on the variant allele and this is driven by the underlying genetic defect. Deciphering the molecular mechanisms by which this happens and its precise timing during embryogenesis requires further studies. But the autosomal dominant Mendelian inheritance pattern of secondary epimutations, with consequently no bias toward a maternal transmission, argues against a specific mechanism and origin of promoter methylation in the oocyte, as hypothesized for primary epimutations.2,11
Among the genetic variants described so far in association with MLH1 epimutation, including the ones reported in this study, some cause complete allelic inactivation by themselves, independently of epigenetic silencing, and promoter methylation may appear as a redundant mechanism of inactivation. The c.-27C>A variant significantly reduces MLH1 transcriptional activity, as demonstrated in reporter assay, and methylation on the c.[-27C>A;85G>T] variant allele can act as a stabilizing mechanism of transcriptional silencing.9 This supports the statement that silencing precedes DNA methylation and that an allele less highly expressed is prone to methylation.37 We demonstrated a functional impact of the genetic alteration by itself in families 1 and 2. In these cases, the assumption can be made that methylation prevents cells from transcription of a nonfunctional allele. However, when testing in our lab other germ-line variants identified within MLH1 5′-UTR, exon 1, and intron 1, no promoter hypermethylation associated with these variants was detected, even for c.116+1G>C and c.116+2T>C, which have validated functional impact on splicing (data not shown). Nucleotide changes at the 5′ end of MLH1 are not systematically associated with promoter methylation, even those showing reduced promoter activity in functional assays.21 These findings support sequence-specific recruitment of epigenetic modifiers or sequence-specific loss of binding of a chromatin insulator that normally prevents MLH1 promoter from epigenetic modification.
Considerable variations in the promoter methylation levels (i.e., very different proportions of PBLs carrying the epimutation) have been observed between patients, even from same family. No correlation has been established so far between the methylation level and individual cancer risk or the age of cancer onset.8 Accordingly, we observed low methylation levels in family 4, and a significant family history of Lynch-type cancers that included several CRC below 50 and multiple CRC in one family member with quasi-undetectable methylation in PBLs. This study supports the fact that epimutation carriers should be considered at high risk of developing cancer irrespective of the methylation level in their PBLs. It also corroborates the requirement for very sensitive methylation screening techniques to identify mosaic epimutations.2,13
In family 1, we observed a tendency toward increasing methylation level over generations. Even if evidence supports the stability of the methylation level throughout life,38 variable clonal expansion of methylated and unmethylated PBLs, as well as a decrease with age in the number of PBLs with a methylated version of the variant allele in some families, cannot be ruled out. Part of the explanation for the increasing methylation level over generations could also lie in the genetic basis of the secondary epimutation, because resistance to demethylation during epigenetic reprogramming has been demonstrated for transposable elements in mice.39
Our long-range PCR NGS strategy successfully identified the exonic insertion of an Alu sequence and enabled extensive sequence variation screening of MLH1 and surrounding regions. This strategy can potentially detect structural variants smaller than 10 kb, provided that they are located within the same amplicon (otherwise allele drop-out will prevent sequencing of the breakpoints). Consequently MLPA technology can still be required to detect some rearrangements.
In conclusion, we present here the largest cohort of patients with a broad spectrum of genetic defects associated with MLH1 promoter methylation. This study illustrates the diversity and complexity of secondary epimutations and the need for additional studies to further explore the molecular mechanisms leading to MLH1 promoter methylation.
Members of the GGC consortium
Caroline Abadie, Emmanuelle Barouk-Simonet, Françoise Bonnet, Chrystelle Colas, Jean-Pierre Fricker, Pascaline Gaildrat, Paul Gesta, Sophie Grandjouan, David Malka, Sylviane Olschwang, Cornel Popovici, Julie Tinat, and Hélène Zattara.
References
Chan TL, Yuen ST, Kong CK et al. Heritable germline epimutation of MSH2 in a family with hereditary nonpolyposis colorectal cancer. Nat Genet 2006;38:1178–1183.
Hitchins MP. The role of epigenetics in Lynch syndrome. Fam Cancer 2013;12:189–205.
Gazzoli I, Loda M, Garber J, Syngal S & Kolodner RD. A hereditary nonpolyposis colorectal carcinoma case associated with hypermethylation of the MLH1 gene in normal tissue and loss of heterozygosity of the unmethylated allele in the resulting microsatellite instability-high tumor. Cancer Res 2002;62:3925–3928.
Tufarelli C, Stanley JA, Garrick D et al. Transcription of antisense RNA leading to gene silencing and methylation as a novel cause of human genetic disease. Nat Genet 2003;34:157–165.
Hansmann T, Pliushch G, Leubner M et al. Constitutive promoter methylation of BRCA1 and RAD51C in patients with familial ovarian cancer and early-onset sporadic breast cancer. Hum Mol Genet 2012;21:4669–4679.
Morgan HD, Santos F, Green K, Dean W & Reik W. Epigenetic reprogramming in mammals. Hum Mol Genet 2005;(14 spec no 1):R47–58.
Crepin M, Dieu MC, Lejeune S et al. Evidence of constitutional MLH1 epimutation associated to transgenerational inheritance of cancer susceptibility. Hum Mutat 2012;33:180–188.
Morak M, Schackert HK, Rahner N et al. Further evidence for heritability of an epimutation in one of 12 cases with MLH1 promoter methylation in blood cells clinically displaying HNPCC. Eur J Hum Genet 2008;16:804–811.
Hitchins MP, Rapkins RW, Kwok CT et al. Dominantly inherited constitutional epigenetic silencing of MLH1 in a cancer-affected family is linked to a single nucleotide variant within the 5′UTR. Cancer Cell 2011;20:200–213.
Barbour VM, Tufarelli C, Sharpe JA et al. Alpha-thalassemia resulting from a negative chromosomal position effect. Blood 2000;96:800–807.
Hitchins MP, Wong JJ, Suthers G et al. Inheritance of a cancer-associated MLH1 germ-line epimutation. N Engl J Med 2007;356:697–705.
Kuiper RP, Vissers LE, Venkatachalam R et al. Recurrence and variability of germline EPCAM deletions in Lynch syndrome. Hum Mutat 2011;32:407–414.
Hitchins MP. Finding the needle in a haystack: identification of cases of Lynch syndrome with MLH1 epimutation. Fam Cancer 2016;15:413–422.
Lynch HT & de la Chapelle A. Hereditary colorectal cancer. N Engl J Med 2003;348:919–932.
Ligtenberg MJ, Kuiper RP, Chan TL et al. Heritable somatic methylation and inactivation of MSH2 in families with Lynch syndrome due to deletion of the 3′ exons of TACSTD1. Nat Genet 2009;41:112–117.
Gylling A, Ridanpaa M, Vierimaa O et al. Large genomic rearrangements and germline epimutations in Lynch syndrome. Int J Cancer 2009;124:2333–2340.
Morak M, Koehler U, Schackert HK et al. Biallelic MLH1 SNP cDNA expression or constitutional promoter methylation can hide genomic rearrangements causing Lynch syndrome. J Med Genet 2011;48:513–519.
Cini G, Carnevali I, Quaia M et al. Concomitant mutation and epimutation of the MLH1 gene in a Lynch syndrome family. Carcinogenesis 2015;36:452–458.
Deng G, Chen A, Hong J, Chae HS & Kim YS. Methylation of CpG in a small region of the hMLH1 promoter invariably correlates with the absence of gene expression. Cancer Res 1999;59:2029–2033.
Kim S, Cho CS, Han K & Lee J. Structural variation of Alu element and human disease. Genomics Inform 2016;14:70–77.
Ward RL, Dobbins T, Lindor NM, Rapkins RW & Hitchins MP. Identification of constitutional MLH1 epimutations and promoter variants in colorectal cancer patients from the Colon Cancer Family Registry. Genet Med 2013;15:25–35.
Kwok CT, Vogelaar IP, van Zelst-Stams WA et al. The MLH1 c.-27C>A and c.85G>T variants are linked to dominantly inherited MLH1 epimutation and are borne on a European ancestral haplotype. Eur J Hum Genet 2014;22:617–624.
Hancks DC & Kazazian HH Jr.. Active human retrotransposons: variation and disease. Curr Opin Genet Dev 2012;22:191–203.
Taskesen M, Collin GB, Evsikov AV et al. Novel Alu retrotransposon insertion leading to Alstrom syndrome. Hum Genet 2012;131:407–413.
Crivelli L, Bubien V, Jones N et al. Insertion of Alu elements at a PTEN hotspot in Cowden syndrome. Eur J Hum Genet 2017;25:1087–1091.
Wimmer K, Callens T, Wernstedt A & Messiaen L. The NF1 gene contains hotspots for L1 endonuclease-dependent de novo insertion. PLoS Genet 2011;7:e1002371.
Wagner A, Barrows A, Wijnen JT et al. Molecular analysis of hereditary nonpolyposis colorectal cancer in the United States: high mutation detection rate among clinically selected families and characterization of an American founder genomic deletion of the MSH2 gene. Am J Hum Genet 2003;72:1088–1100.
Liu Q, Hesson LB, Nunez AC et al. A cryptic paracentric inversion of MSH2 exons 2-6 causes Lynch syndrome. Carcinogenesis 2016;37:10–17.
van der Klift H, Wijnen J, Wagner A et al. Molecular characterization of the spectrum of genomic deletions in the mismatch repair genes MSH2, MLH1, MSH6, and PMS2 responsible for hereditary nonpolyposis colorectal cancer (HNPCC). Genes Chromosomes Cancer 2005;44:123–138.
Charbonnier F, Baert-Desurmont S, Liang P et al. The 5′ region of the MSH2 gene involved in hereditary non-polyposis colorectal cancer contains a high density of recombinogenic sequences. Hum Mutat 2005;26:255–261.
Kloor M, Sutter C, Wentzensen N et al. A large MSH2 Alu insertion mutation causes HNPCC in a German kindred. Hum Genet 2004;115:432–438.
Bennett EA, Keller H, Mills RE et al. Active Alu retrotransposons in the human genome. Genome Res 2008;18:1875–1883.
Meissner A, Mikkelsen TS, Gu H et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 2008;454:766–770.
Xiang S, Liu Z, Zhang B et al. Methylation status of individual CpG sites within Alu elements in the human genome and Alu hypomethylation in gastric carcinomas. BMC Cancer 2010;10:44.
Wang X, Fan J, Liu D, Fu S, Ingvarsson S & Chen H. Spreading of Alu methylation to the promoter of the MLH1 gene in gastrointestinal cancer. PLoS One 2011;6:e25913.
Venkatachalam R, Ligtenberg MJ, Hoogerbrugge N et al. Germline epigenetic silencing of the tumor suppressor gene PTPRJ in early-onset familial colorectal cancer. Gastroenterology 2010;139:2221–2224.
Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 2012;13:484–492.
Sloane MA, Nunez AC, Packham D et al. Mosaic epigenetic inheritance as a cause of early-onset colorectal cancer. JAMA Oncol 2015;1:953–957.
Popp C, Dean W, Feng S et al. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature 2010;463:1101–1105.
Acknowledgments
This work was supported by Lille University Hospital (Fonds Hospitalier d’Aide à l’Emergence et la Structuration des Activités et Équipes de Recherche).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Disclosure
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Leclerc, J., Flament, C., Lovecchio, T. et al. Diversity of genetic events associated with MLH1 promoter methylation in Lynch syndrome families with heritable constitutional epimutation. Genet Med 20, 1589–1599 (2018). https://doi.org/10.1038/gim.2018.47
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gim.2018.47
Keywords
This article is cited by
-
Identifying primary and secondary MLH1 epimutation carriers displaying low-level constitutional MLH1 methylation using droplet digital PCR and genome-wide DNA methylation profiling of colorectal cancers
Clinical Epigenetics (2023)
-
Massive parallel sequencing in individuals with multiple primary tumours reveals the benefit of re-analysis
Hereditary Cancer in Clinical Practice (2021)
-
Highly sensitive MLH1 methylation analysis in blood identifies a cancer patient with low-level mosaic MLH1 epimutation
Clinical Epigenetics (2019)