Abstract
The introduction of diagnostic clinical genome and exome sequencing (CGES) is changing the scope of practice for clinical geneticists. Many large institutions are making a significant investment in infrastructure and technology, allowing clinicians to access CGES, especially as health-care coverage begins to extend to clinically indicated genomic sequencing-based tests. Translating and realizing the comprehensive clinical benefits of genomic medicine remain a key challenge for the current and future care of patients. With the increasing application of CGES, it is necessary for geneticists and other health-care providers to understand its benefits and limitations in order to interpret the clinical relevance of genomic variants identified in the context of health and disease. New, collaborative working relationships with specialists across diverse disciplines (e.g., clinicians, laboratorians, bioinformaticians) will undoubtedly be key attributes of the future practice of clinical genetics and may serve as an example for other specialties in medicine. These new skills and relationships will also inform the development of the future model of clinical genetics training curricula. To address the evolving role of the clinical geneticist in the rapidly changing climate of genomic medicine, two Clinical Genetics Think Tank meetings were held that brought together physicians, laboratorians, scientists, genetic counselors, trainees, and patients with experience in clinical genetics, genetic diagnostics, and genetics education. This article provides recommendations that will guide the integration of genomics into clinical practice.
Genet Med 18 11, 1075–1084.
Similar content being viewed by others
Main
Genetic diagnostics has evolved rapidly over the past six decades, changing the practice of clinical genetics and helping all medical specialties to achieve improved clinical outcomes. Techniques to see the correct complement of human chromosomes1 led to genomic diagnostics through light microscopy. Three years later, the first disorder identified as being genomic, Down syndrome, was found to be associated with trisomy 21 (ref. 2). For the next 30 years, the application of this “genomic” technology was limited to diagnosing various germ-line and somatic aneuploidies and translocations, with increasing resolution and, beginning in the late 1980s, the use of fluorescence in situ hybridization.3,4,5 Comparative genomic hybridization and single-nucleotide polymorphism array–based methodologies enabled the first genome-wide high-resolution analysis for small deletions and duplications6 and replaced the G-banded karyotype as a first-tier diagnostic test.7 Ten years later, genome sequencing8 and exome sequencing9,10,11,12 were first applied in clinical medicine and have since become widely used in diagnostics.13
With the expanded comprehensiveness of genetic testing, the interpretation of results has become more complex. Clinical genome and exome sequencing (CGES) promises to improve identification of the molecular determinants for many disease categories, but it carries a degree of uncertainty given the large number of variants of uncertain significance identified in any individual, as well as secondary findings unrelated to the primary indication for testing. Secondary findings are identified in 0.5–3.5% of individuals undergoing CGES,14,15,16,17,18 but they will increase as our knowledge about genetic determinants of disease advances. As CGES evolves toward screening healthy populations, what we now consider secondary findings will become the primary findings, which will form the foundation for the practice of genomic precision medicine.19
The increasing complexity of CGES coupled with the broad-scale application to a wide range of medical conditions requires a transformative approach to the ways in which we currently practice clinical genetics—and medicine in general. New clinical models are needed to enable the integration of clinically relevant genomic information into specialized and routine health care.
Clinical Genetics Think Tank
The concept of a Clinical Genetics Think Tank (CGTT) evolved as it became evident that several clinical genetics centers in the United States and Canada were independently developing research and clinical programs to respond to demands for genomic services and were facing common challenges.20 The goals of the CGTT were to identify the major challenges to integrating CGES into clinical practice and to provide practical recommendations based on published evidence and professional experience.
Here, we outline the recommendations agreed on by the CGTT, presented as a framework to influence policy discussions, offer guidance in the creation of new individualized care models incorporating family and patient preferences, and provide direction regarding how to integrate new genomic knowledge across organizations to ensure optimal management of patients undergoing CGES.
Five critical areas of focus in the clinical workflow to effectively integrate CGES were identified: the pretest process, pretest education for patients and providers, phenotyping, sequence data interpretation, and posttest patient care. For each area, we present challenges and recommendations followed by key questions, summarized in Table 1 .
Pretest Process: Patient Selection and Financial Coverage Issues for CGES
Many of the challenges of integrating CGES into clinical care are directly related to its vast diagnostic potential and its cost.20,21 As the cost/benefit ratio of CGES improves, some barriers to ordering this test are likely to diminish.22 The diagnostic value will increase as genomic and phenotypic data are shared and interpreted, making it imperative that a robust pretest process be developed to enable clinicians to utilize this test in the care of patients most likely to benefit.
Challenges and recommendations
Who should order CGES? The American College of Medical Genetics and Genomics (ACMG) has recommended that CGES be accompanied by consultation with a genetics professional and adequate genetics counseling.23 Given the relatively small number of genetics professionals in the workforce and the projected increase in demand for CGES, the CGTT recommends that a clinician be able to order CGES if he or she has at least a minimum knowledge base and an opportunity to subsequently request additional expertise from a genetics professional (e.g., for pretest evaluation or interpretation).
This minimum knowledge is defined as the ability to do the following:
-
1. Perform a basic clinical genetics evaluation, including family history, genetics-focused medical history, and physical examination, and a critical summary of prior genetic evaluations
-
2. Determine whether CGES is the test of choice for the specific clinical indication, considering other available tests (including genomic and nongenomic tests such as metabolic screening) and the patient’s age and cognitive ability
-
3. Provide adequate pretest counseling, including informed consent for primary and secondary findings
-
4. Interpret the results of the CGES and provide adequate posttest counseling or seek input for this step from a genetics professional
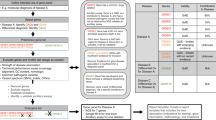
What are the clinical indications to order CGES? There are limited publications guiding the decision to order CGES.13,24,25 Figure 1 shows an algorithm modified from a prior guideline;25 it may require further modifications as evidence is published regarding the sensitivity of CGES in specific patient populations.
Algorithm to help identify patients most likely to benefit from clinical genome and exome sequencing (CGES). Genetic testing should be a phenotype-driven process, taking into account the most current knowledge of the genetic basis of the condition(s) undergoing investigation. The decision to order CGES versus a targeted panel will be informed by the genetic heterogeneity of the condition, depth of coverage for genes of interest, and the cost of each test. *Consider CGES as a first-line test when there is parental consanguinity. FISH, fluorescent in situ hybridization.
The following general principles guide this algorithm:
-
1. When the patient’s phenotype is specific to a known genetic condition for which optimized genetic panel testing exists, the targeted gene panel should be given priority assuming it is more sensitive (e.g., for Noonan spectrum disorders).
-
2. Chromosome microarray analysis should be considered prior to CGES when the phenotype is not specific for a known genetic condition and when the family history does not indicate X-linked or autosomal recessive inheritance. This approach is suggested until the copy-number variation–calling sensitivity of CGES is equivalent to that of chromosome microarray platforms, and it may become obsolete once genome sequencing (GS) is used as a platform, given the potential capability of GS to detect copy-number variation at the same sensitivity as chromosome microarray.26,27
-
3. CGES should be considered prior to chromosome microarray when there is parental consanguinity.28
-
4. CGES should not be considered a mandatory next step when a targeted gene panel does not provide a diagnostic result. However, CGES in silico analysis may initially be focused on a specific subset of genes and subsequently expanded to the full data set if the initial analysis is nondiagnostic (e.g., http://www.genedx.com/test-catalog/xomedxslice/). This approach may become ubiquitous as the cost of CGES falls and diagnostic laboratories streamline their workflows and maintain a “single-platform” genetic test. Until then, clinicians may order CGES after a nondiagnostic gene panel if (i) there remains strong suspicion for a genetic cause and the gene panel previously ordered is outdated, (ii) the patient’s phenotype has evolved or is atypical for the negative panel, or (iii) there is clinical urgency regarding the diagnosis and/or immediate clinical utility for family members who may be at risk.
Should there be an institutional gatekeeper? Some institutions have established a gatekeeper role for access to CGES to promote appropriate patient selection.21 Boston Children’s Hospital BEST (Bringing Excellence to Selecting Tests) committee consists of members with subspecialty expertise who review all requests for CGES and provide educational feedback to the ordering clinician if a request is declined. Applications are initially reviewed by the content expert most closely related to the phenotype, who may approve the application or send it for full committee review. Given the potential benefits of education, quality control, and cost control associated with a gatekeeping role, institutions should give consideration to developing such a process, with diverse representation from institutional stakeholders. The precise model for each institution will vary, but it may include the provision of guidelines (either institution-specific or peer-reviewed publications) that are a prerequisite to accessing CGES in addition to (or instead of) an individual or committee review of each application. The skill set required to review CGES applications is likely to be the same as that required to order and interpret the test, as outlined above under “Who should order CGES?.”
How should insurance approval for CGES be obtained? Insurance companies and the Canadian Provincial Ministries of Health differ in their policies toward covering the cost of CGES. Several insurance providers have specific policies limiting, and in some cases denying, access based on the perception of CGES as an experimental or investigational technology.29 In the absence of access to CGES, less appropriate tests, often of equivalent or increased cost, may be ordered because those tests are covered by insurance.30
A recent study looking at insurance coverage for exome sequencing found extreme variability both among and within insurance providers that was heavily dependent on the indication for testing and often involved large costs to the patients due to high deductibles and copays even when approved.22 A registry of next-generation sequencing tests and reimbursement policies has been set up at the University of California–San Francisco’s Center for Translational and Policy Research on Personalized Medicine (https://pharm.ucsf.edu/transpers) to track and inform decisions made by insurance representatives. Genetics professionals spend significant time writing letters of medical necessity to justify testing, and patients are referred to clinical genetics services for assistance in this process. Increased demand for this type of assistance would limit patient access to CGES because clinical genetics services are not available in the majority of health-care settings. In the short term, sample letters of medical necessity should be made publically available to provide ordering clinicians the opportunity to adapt appropriately worded letters for their specific patient request. The letters should include references to regularly updated, peer-reviewed literature on clinical validity and utility and patient satisfaction associated with CGES.
In the longer term, enhanced communication between professional bodies (advocating for CGES to be used on the basis of published evidence for improved diagnostic, management, and patient-satisfaction outcomes) and payers (articulating the industry’s prerequisites to providing coverage for CGES) is necessary to increase accessibility to CGES. The American Society of Human Genetics, ACMG, and Canadian College of Medical Geneticists groups will need to play an active role and continue to work with payers to substantiate the clinical validity and utility of CGES. Periodic publication of practice guidelines by these professional bodies should be part of this effort.24
Pretest Education for Patients and Providers
Multiple stakeholders require education about CGES, including physicians, genetics counselors, trainees, insurance and government stakeholders, laboratorians, and patients and their families. Education and informed consent have been thoroughly discussed elsewhere.31,32,33,34,35,36,37 However, to facilitate broad implementation of CGES into clinical care, some areas require refinement in the context of increasingly advanced and complex genomic testing.
Challenges and recommendations
Who requires education, and what are the key topics?
Patients. Patient education regarding CGES should be designed specifically for the target audience and extend beyond written information in the informed-consent document. Several published articles and guidelines have outlined the topics considered essential to the informed-consent process.38,39 The CGTT, primed by its patient and parental participants, concluded that it is most important to focus education on the types of results that are possible. The key patient-education topics identified by the CGTT are summarized in Table 2 .
Providers. Provider education should be designed to enable assessment by clinicians of their ability and educational needs prior to ordering CGES. Topics should include the following: what a basic clinical genetics evaluation and family history entails, review of different types of genetic tests to allow a critical summary of prior evaluations, overview of CGES (indications, strengths, and limitations) to enable clinicians to determine whether this is the best test for the specific clinical indication, what pretest counseling involves, interpretation of results, posttest counseling, and when to seek support from a genetics professional.
Insurance and government stakeholders. Insurance providers and government bodies require an understanding of CGES to make informed decisions about test coverage and reimbursement levels. Key education topics are the clinical validity as it relates to the diagnosis and management of patients and their families as well as the utility of CGES for specific patient populations, which will be informed by data from research that is largely in its infancy.
How is education provided? Clinician–patient interaction is key for adequate patient education; however, patient-education materials should be provided as a supplement to the patient–clinician interaction.40 Numerous resources appropriate for patient education in CGES are available online at no cost (see Appendix). Their formats include video, interactive modules, and expert-authored test descriptions designed to meet different educational requirements. Some have been translated into multiple languages to minimize disparities in access to CGES.
Clinical geneticists are already highly competent in many of the areas required to order and interpret CGES; however, obtaining the specific skills required for clinical interpretation of CGES is a critical element of translating sequence into patient care. Development of specialty-specific educational opportunities should be promoted, such as the genomics case conferences sponsored by ACMG that focus on variant interpretation as it relates to CGES. Other short courses and continuing medical education (CME) modules are available across North America and worldwide, and details can be found on the websites of the professional societies including the American Society of Human Genetics (http://www.ashg.org/education/Health_Professionals.shtml), ACMG (http://www.acmg.net/ACMG/Education/), and the European Society of Human Genetics (http://www.eshg.org/courses.0.html). Clinician education methods may also include “on-demand” learning; for example, the electronic health record may contain prompts to training material when CGES is being electronically ordered.
Phenotyping: Collecting Accurate, Standardized Phenotype Data to Assist in Test Interpretation
The integration of clinical data, particularly a patient’s phenotype, is of central importance in determining what variants are most relevant during CGES interpretation. Accurate and consistent phenotype measures are needed to fully realize the utility of CGES; however, phenotype data are obtained by a wide variety of medical specialties across many health-care systems, and current methods for determining and recording phenotype information are not standardized. Recognition of the need to use standardized terms and measures for optimal phenotyping has stimulated worthy educational efforts to define phenotypic terms.41,42,43,44,45,46,47,48,49 One powerful approach to standardizing clinical data is the Human Phenotyping Ontology project (http://www.human-phenotype-ontology.org). The project, which was designed to reflect normal clinical phenotyping processes, is “a computational representation of a domain of knowledge (phenotype) based on a controlled, standardized vocabulary for describing entities and the semantic relationships between them” (ref. 50; see also refs. 51 and 52). Collecting clinical data for standardized phenotyping requires clinical databases to be either created or modified from preexisting tools such as Phenotips53 (http://phenotips.org). This resource supports the use of Human Phenotyping Ontology project terms and allows for data-entry functionality for PhenomeCentral or PhenoDB (http://phenodb.net), giving clinicians secure access to genotype–phenotype information and enabling them to collaborate on diagnoses. A full summary of online phenotype–genotype resources can be found in Supplementary Table S1 online.
One approach to maximizing the utility of standardized phenotypic data for analyzing CGES data would be to grant laboratory interpreting staff access to phenotypic data.
Challenges and recommendations
How can consistent and uniform phenotype data be collected? The phenotype may be focused for a specific clinical condition, suggesting a modular approach to phenotyping with the data content of individual modules defined for specific organ systems or diseases. In addition, a universal phenotypic data set could be established to assist in interpretation of as many primary and secondary findings on genomic testing as possible. Phenotypic measures developed for disease-specific research purposes (e.g., obesity) will help define which data should be captured for optimal CGES interpretation. Electronic health records (EHRs) need to be adapted to encompass universally consistent ontologic terms allowing for a common phenotyping language, which would play an essential role in complete annotation of the variants in the human exome and genome from a clinical perspective.
Once phenotypic fields are delineated, it will be critical to get buy-in across organizations, nationally and internationally, to help disseminate robust information related to the significance of variants and mutations and their impact on phenotype, management, and clinical outcomes. A centralized and well-curated database containing both clinical and research genomic results linked to standardized phenotypic data would rapidly increase the accuracy of variant interpretation. Large-scale data collection will require broad-scale efforts and institutional support, but the benefits in terms of standardized care, portability of information, and improved metrics to inform cost/benefit analyses would be potentially great.
How can dynamic phenotypes be captured in order to refine variant interpretation? The ability to update phenotypic fields will be essential to understand the significance of variants identified on genomic testing across an individual’s life span. There is also a need to reassess variants of uncertain significance that may have been implicated in human disease in the interval since testing was performed. A post-CGES follow-up visit with a genetics professional would provide an opportunity to proactively manage patients in relation to their identified disease and risk variants, as well as update patients with new information related to the variants identified on initial testing. These visits would also provide an opportune time to update and establish comprehensive phenotypes on individuals and to institute the application of correct terminologies into the EHR. Establishment of automated care assistant models that could link genomic variants to populations at risk and provide recommended management options would streamline the process of reviewing an individual’s genome. Initial work in this domain is actively being pursued by research consortia supported through the NIH, including the Electronic Medical Records and Genomics (eMERGE) Network (https://emerge.mc.vanderbilt.edu), the Clinical Sequencing Research Consortium (CSER) (https://cser-consortium.org), and the Implementing Genomics in Practice (IGNITE) consortium (http://www.ignite-genomics.org/ignite_about.html). Recent work in applying such models to pharmacogenetic variants has shown the potential viability of such an approach.54,55
Sequence Data Interpretation: Managing the Integration of Clinical and Genomic Information to Generate a Comprehensive Clinical Report
Much of the workflow (sequencing, alignment, variant calling) is standardized in diagnostic genomics laboratories. In comparison with gene panel testing, CGES interpretation requires closer collaboration between laboratorians and clinicians in order to reduce the genomic variants to be analyzed to a reasonable number. Fostering existing relationships and processes and establishing new ones to support these important interactions will ultimately ensure that CGES interpretation is accurate and efficient and that the relevant information is communicated in a comprehensive clinical report.
Challenges and recommendations
How should laboratorians and clinicians interact to determine whether CGES is the appropriate test for a specific patient? Determination of the correct diagnostic test for an individual patient should be the decision of the physician caring for the patient; however, the diagnostic laboratory directors and genetics counselors should be available to provide feedback regarding the suitability of CGES relative to the clinical indication. Factors such as how well relevant genes are covered by CGES as well as what types of variation are predominant in those genes are important for the ordering clinician to understand (e.g., triplet repeat expansions, copy-number variants, and other structural variants may not be well covered by CGES).
How should variants be prioritized? Identification and annotation of variants that are sufficiently relevant to the patient’s phenotype to be included in a focused clinical report are complex steps in the process of CGES interpretation.54,55,56,57 Maintaining high stringency with respect to proof of pathogenicity at both gene and variant levels will help to reduce the potentially severe consequences of misdiagnosis.58 The following recommendations aim to optimize this process:
-
1. Where possible, laboratories should establish gene lists that relate to specific clinical indications for CGES to help streamline initial variant calling and annotation. Because population frequency resources are better integrated into pipelines reducing the number of variants to assess, the need to define gene lists in advance may be less relevant. Establishing gene lists does not imply that laboratories should limit CGES analysis to specific gene sets; however, lists of genes known to be relevant to the patient’s indication for testing are helpful for prioritizing variants for deeper review and for determining effective coverage. Community efforts to establish the level of evidence for gene–disease relationships, such as those undertaken by ClinGen (http://www.clinicalgenome.org/knowledge-curation/gene-curation/), will be critical in assessing which genes to review and how to report variants in those genes. These lists will require continual review and updating.
-
2. Each laboratory should establish a comprehensive database to track their variant interpretations and evidence for classification to enable this information to be shared and compared with other laboratories through publically available resources such as the ClinVar database.59
Should there be laboratory standards relating to depth of coverage and variant validation methods? Variability in depth of coverage across the exome and genome leads to the possibility of gaps in sequencing and missing or uninterpretable data due to regions with high homology. The laboratory should make clear whether it will ensure completion or “fill-in” of any missing sequences or classes of variation that are relevant to the patient’s indication but technically difficult to access by CGES.55 Currently, laboratories are not routinely “filling in” gaps with Sanger sequencing. Diagnostic laboratories should establish mechanisms by which clinicians can be informed of technical test limitations on a patient-specific and indication-specific basis.
The common practice of confirming all pathogenic variants included in a clinical report by Sanger sequencing will become less warranted as validation efforts increase over time and as laboratories ensure integrity of the sample identity across complex genomic sequencing workflows.
Which variants should be included in the clinical report? A critical issue is the decision regarding which variants are included in a formal clinical report.60 Results should be reported that definitively or likely explain the indication for CGES. If the laboratory decides to report variants in genes of uncertain significance (GUS), such as those with limited, no, or conflicting evidence according to ClinGen’s framework but that represent strong candidates to explain the clinical findings, then efforts should be made to refer to research-based evaluation for further clarification; it should be made explicit when evidence is lacking. Clinicians and CGES laboratory staff should be cognizant of the distinction between research and clinical care. Deposition of cases with candidate genes into Matchmaker Exchange is recommended (http://www.matchmakerexchange.org/). For secondary findings, the threshold for inclusion in a clinical report should be more stringent than for the primary indication, and a minimal standard set of genes for inclusion of secondary variants (e.g., the ACMG recommendations)61 should be considered. The types of variants reported should be limited to likely pathogenic or pathogenic.54 In addition, reports should make it clear that complete detection and interpretation of all disease-associated variants is not possible.
Posttest Patient Care: Return of Primary and Secondary Findings and Reevaluation of CGES Data
The posttest patient-care phase begins once a CGES report has been returned to the ordering physician. In theory, this phase could last a lifetime because the aspirational goal is to use DNA sequencing results to make a primary genomic diagnosis and to maintain health for a generation and even beyond. Additional factors that support the notion of a continuum of posttest patient care are the ever-changing methods of DNA analysis, the growing body of evidence and rules for interpreting the pathogenic potential of a variant, the anticipated inclusion of “omics” methods, and animal models to further delineate clinical phenotypes. The initial appointment to discuss CGES results is likely to be the first of several posttest contacts between the patient and the medical team.
Challenges and recommendations
How should primary and secondary results be returned? The process of returning CGES results to the patient and other members of the medical team must be tailored to the diagnostic issue, the patient and family needs, the clinical setting, and the available workforce. The urgency and complexity of information to be returned to the patient should be balanced according to the patient’s stated wishes obtained during the pretest consent process. Patients should have the choice of one or more results disclosure sessions to improve understanding and to avoid fatigue. Although we understand that it may be difficult to accommodate two sessions, and that in many circumstances only one session may be preferable for families, the feedback received from families and patient advocates should challenge the current health-care model to allow for more than one session to discuss results of CGES. The first session addresses the principal diagnostic result. If the diagnostic or management issues relating to the primary diagnosis are outside the expertise of the ordering clinician, then additional medical specialists may be invited to attend. Alternatively, the patient should be made aware of the need for obtaining specialist opinion(s), and the ordering clinician would complete a comprehensive referral including interpretation of the CGES results.
The second session should address secondary findings that are medically actionable and should be offered within a short time period after the first session if such findings exist. Pharmacogenomics variants would be addressed at this time in consultation with a pharmacist or pharmacologist trained in interpreting pharmacogenetic data. This process is summarized in Figure 2 .
Integration of CGES into EHRs A significant number of institutions have implemented large-scale EHR systems that are uniformly aimed at improving patient care and safety, and also address billing-related issues. As CGES moves into daily clinical care, several providers of EHRs have begun developing modules that allow full integration of CGES results into the EHR of each patient. Several aspects must be considered for this to occur; these are beyond the scope of this article. However, it is important to emphasize that detailed phenotypic analyses accompanied by primary, secondary, and pharmacogenetic findings resulting from CGES should form part of the EHR in the most transparent way to ensure patient safety and appropriate management.
Should genomic reevaluation be offered? Periodic reevaluation of an individual’s genome with the aim of refining diagnostic interpretation is desirable but it requires significant time and financial commitments from laboratories, clinicians, and patients. Reevaluation would apply both to individuals whose CGES was diagnostic for their primary disorder and to those who did not receive a genomic diagnosis. A major issue concerning reevaluation of CGES clinically is how to bill and be reimbursed for these services. The value of reevaluation has been demonstrated in the research realm, and several initiatives have demonstrated that automated requerying of CGES data to capture novel disease-associated variants is computationally possible. These initiatives are also able to compare variants among related cohorts to identify novel pathogenic variants associated with specific phenotypes. An excellent example of this is the Epilepsy Genetics Initiative (http://www.cureepilepsy.org/egi/index.asp), which is warehousing phenotype and genotype data (primarily exomes) contributed by clinicians and investigators and offering a reanalysis of the entire cohort every 6 months. Although several diagnostic laboratories are offering free reanalysis within a specified time frame, guidelines need to be set for reimbursement for the diagnostic laboratories for these types of labor-intensive services. With 50–80% of clinical CGES tests having no identifiable definitive pathogenic variant on initial analysis, subsequent reanalysis as new information and pathogenic variants are identified will be critical. Ideally, at routine follow-up appointments, the family and personal health history would be updated, original sequencing results and interpretation would be reviewed, and DNA variant databases would be requeried. If indicated, the physical examination would be repeated and follow-up tests ordered. As functional testing becomes more accessible, variants of uncertain significance may be reclassified to allow diagnostic certainty. Supplementary “omics” profiles, including metabolomics, transcriptomics, epigenomics, and metagenomics, as well as animal models, already widely used in research may enable functional correlation with important genomic variants. These innovations should be the subject of early research on costs and effectiveness.
Conclusion
Articles have been published that outline issues related to implementation of genomic medicine into the clinic. Some focus on specific issues such as the evolving role of the medical geneticist,62 optimization and integration of genomic information in the EHR,63 research needs,64 ethical and legal aspects of returning results and distinguishing research from clinical care,65 and descriptions of institutional approaches to integration of genomic information into clinical care.20,66,67 Few have dealt with the practical aspects of implementation of genomic medicine into the clinic. The review by Biesecker and Green13 provided an overview of CGES, but it did not focus on the obstacles to implementation. At a 2011 Colloquium of Genomic Medicine, sponsored by the US National Human Genome Research Institute, 20 groups of clinicians and investigators working on projects to implement genomic medicine summarized early findings.21 Some common challenges were seen: lack of consensus on the pathogenicity and clinical relevance of identified variants, reimbursement for genomic tests and interventions, the burden on clinicians and patients in managing the information generated from genomic testing, and infrastructural needs, such as an open-access knowledge base for cataloging variants and phenotypes. The colloquium recognized that much of the work in clinical genomic medicine was being done in isolation and would benefit from active collaborative efforts and sharing of best practices, a catalyst for the CGTT workshops and this article.
We hope that this outline will serve as the basis for continued interactions of various professional groups and facilitate the establishment of collaborative infrastructures that are essential to implement best practices in clinical genomic medicine. It will only be through collaborative efforts and the application of consensus recommendations from different institutions and diverse populations (ethnically and racially, economically, and prenatal, pediatric, and adult, for a wide spectrum of disease states and in healthy populations) that the robust data and metrics needed to evaluate the impact of genomic medicine on the health of individuals and populations will be able to be evaluated and best practice guidelines established for implementation of genomic medicine.
Disclosure
The authors declare no conflict of interest.
Appendix: Educational resources
Accessed 8 April, 2015
1. http://www.genome.gov/glossary/index.cfm?
2. https://www.my46.org/learning-center
3. https://familyhistory.hhs.gov/fhh-web/home.action
4. http://www.geneticseducation.nhs.uk/
5. http://www.cdc.gov/genomics/default.htm
6. http://genes-r-us.uthscsa.edu/resources/genetics/primary_care.htm
7. http://www.kumc.edu/gec/geneinfo.html
8. http://learn.genetics.utah.edu/
9. http://www.accessexcellence.org/AE/AEPC/NIH/index.php
10. http://www.yourgenome.org/landing_dgg.shtml
11. http://www.sickkids.ca/CGM/education/resources/education-resources.html
References
Tjio JH, Levan A. The chromosome number of man. Hereditas 1956;42:1–6. http://onlinelibrary.wiley.com/doi/10.1111/j.1601-5223.1956.tb03010.x/abstract.
Lejeune J, Gautier M, Turpin R. [Study of somatic chromosomes from 9 mongoloid children]. C R Hebd Seances Acad Sci 1959;248:1721–1722.
Pinkel D, Gray JW, Trask B, van den Engh G, Fuscoe J, van Dekken H. Cytogenetic analysis by in situ hybridization with fluorescently labeled nucleic acid probes. Cold Spring Harb Symp Quant Biol 1986;51 Pt 1:151–157.
Kølvraa S, Koch J, Gregersen N, et al. Application of fluorescence in situ hybridization techniques in clinical genetics: use of two alphoid repeat probes detecting the centromeres of chromosomes 13 and 21 or chromosomes 14 and 22, respectively. Clin Genet 1991;39:278–286.
Magenis RE, Maslen CL, Smith L, Allen L, Sakai LY. Localization of the fibrillin (FBN) gene to chromosome 15, band q21.1. Genomics 1991;11:346–351.
Pinkel D, Segraves R, Sudar D, et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat Genet 1998;20:207–211.
Miller DT, Adam MP, Aradhya S, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet 2010;86:749–764.
Ley TJ, Mardis ER, Ding L, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature 2008;456:66–72.
Ng SB, Buckingham KJ, Lee C, et al. Exome sequencing identifies the cause of a mendelian disorder. Nat Genet 2010;42:30–35.
Choi M, Scholl UI, Ji W, et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc Natl Acad Sci USA 2009;106:19096–19101.
Ng SB, Turner EH, Robertson PD, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature 2009;461:272–276.
Maher B. Exome sequencing takes centre stage in cancer profiling. Nature 2009;459:146–147.
Biesecker LG, Green RC. Diagnostic clinical genome and exome sequencing. N Engl J Med 2014;370:2418–2425.
Johnston JJ, Rubinstein WS, Facio FM, et al. Secondary variants in individuals undergoing exome sequencing: screening of 572 individuals identifies high-penetrance mutations in cancer-susceptibility genes. Am J Hum Genet 2012;91:97–108.
Dorschner MO, Amendola LM, Turner EH, et al.; National Heart, Lung, and Blood Institute Grand Opportunity Exome Sequencing Project. Actionable, pathogenic incidental findings in 1,000 participants’ exomes. Am J Hum Genet 2013;93:631–640.
Tabor HK, Auer PL, Jamal SM, et al.; NHLBI Exome Sequencing Project. Pathogenic variants for Mendelian and complex traits in exomes of 6,517 European and African Americans: implications for the return of incidental results. Am J Hum Genet 2014;95:183–193.
Amendola LM, Dorschner MO, Robertson PD, et al. Actionable exomic incidental findings in 6503 participants: challenges of variant classification. Genome Res 2015;25:305–315.
Jurgens J, Ling H, Hetrick K, et al. Assessment of incidental findings in 232 whole-exome sequences from the Baylor-Hopkins Center for Mendelian Genomics. Genet Med 2015;17:782–788.
Biesecker LG. Incidental variants are critical for genomics. Am J Hum Genet 2013;92:648–651.
Bowdin S, Ray PN, Cohn RD, Meyn MS. The genome clinic: a multidisciplinary approach to assessing the opportunities and challenges of integrating genomic analysis into clinical care. Hum Mutat 2014;35:513–519.
Manolio TA, Chisholm RL, Ozenberger B, et al. Implementing genomic medicine in the clinic: the future is here. Genet Med 2013;15:258–267.
Atwal PS, Brennan ML, Cox R, et al. Clinical whole-exome sequencing: are we there yet? Genet Med 2014;16:717–719.
Points to consider in the clinical application of genomic sequencing. Genet Med 2012;14:759–761.
Boycott K, Hartley T, Adam S, et al.; Canadian College of Medical Geneticists. The clinical application of genome-wide sequencing for monogenic diseases in Canada: Position Statement of the Canadian College of Medical Geneticists. J Med Genet 2015;52:431–437.
Shashi V, McConkie-Rosell A, Rosell B, et al. The utility of the traditional medical genetics diagnostic evaluation in the context of next-generation sequencing for undiagnosed genetic disorders. Genet Med 2014;16:176–182.
Gilissen C, Hehir-Kwa JY, Thung DT, et al. Genome sequencing identifies major causes of severe intellectual disability. Nature 2014;511:344–347.
Yuen RK, Thiruvahindrapuram B, Merico D, et al. Whole-genome sequencing of quartet families with autism spectrum disorder. Nat Med 2015;21:185–191.
Teeuw M, Waisfisz Q, Zwijnenburg PJ, et al. First steps in exploring prospective exome sequencing of consanguineous couples. Eur J Med Genet 2014;57:613–616.
Deverka PA, Kaufman D, McGuire AL. Overcoming the reimbursement barriers for clinical sequencing. JAMA 2014;312:1857–1858.
Chakradhar S. Insurance companies are slow to cover next-generation sequencing. Nat Med 2015;21:204–205.
Vrijenhoek T, Kraaijeveld K, Elferink M, et al. Next-generation sequencing-based genome diagnostics across clinical genetics centers: implementation choices and their effects. Eur J Hum Genet 2015;23:1142–1150.
Scollon S, Bergstrom K, Kerstein RA, et al. Obtaining informed consent for clinical tumor and germline exome sequencing of newly diagnosed childhood cancer patients. Genome Med 2014;6:69.
Henderson GE, Wolf SM, Kuczynski KJ, et al. The challenge of informed consent and return of results in translational genomics: empirical analysis and recommendations. J Law Med Ethics 2014;42:344–355.
Bergner AL, Bollinger J, Raraigh KS, et al. Informed consent for exome sequencing research in families with genetic disease: the emerging issue of incidental findings. Am J Med Genet A 2014;164A:2745–2752.
Appelbaum PS, Parens E, Waldman CR, et al. Models of consent to return of incidental findings in genomic research. Hastings Cent Rep 2014;44:22–32.
Points to consider for informed consent for genome/exome sequencing. Genet Med 2013;15:748–749.
Levenseller BL, Soucier DJ, Miller VA, Harris D, Conway L, Bernhardt BA. Stakeholders’ opinions on the implementation of pediatric whole exome sequencing: implications for informed consent. J Genet Couns 2014;23:552–565.
Ayuso C, Millán JM, Mancheño M, Dal-Ré R. Informed consent for whole-genome sequencing studies in the clinical setting. Proposed recommendations on essential content and process. Eur J Hum Genet 2013;21:1054–1059.
Tabor HK, Stock J, Brazg T, et al. Informed consent for whole genome sequencing: a qualitative analysis of participant expectations and perceptions of risks, benefits, and harms. Am J Med Genet A 2012;158A:1310–1319.
National Coalition for Health Professional Education in Genetics. Core competencies in genetics for health professionals. 2007. http://www.nchpeg.org/index.php?option=com_content&view=article&id=237.
Hennekam RC, Allanson JE, Biesecker LG, Carey JC, Opitz JM, Vilain E. Elements of morphology: standard terminology for the external genitalia. Am J Med Genet A 2013;161A:1238–1263.
Carey JC. Editor’s forward to special articles: elements of morphology. Am J Med Genet A 2013;161A:2710.
Biesecker LG, Carey JC. Implementing the elements of morphology in the American Journal of Medical Genetics. Am J Med Genet A 2011;155A:969–971.
Hennekam RC, Cormier-Daire V, Hall JG, Méhes K, Patton M, Stevenson RE. Elements of morphology: standard terminology for the nose and philtrum. Am J Med Genet A 2009;149A:61–76.
Hunter A, Frias JL, Gillessen-Kaesbach G, Hughes H, Jones KL, Wilson L. Elements of morphology: standard terminology for the ear. Am J Med Genet A 2009;149A:40–60.
Allanson JE, Cunniff C, Hoyme HE, McGaughran J, Muenke M, Neri G. Elements of morphology: standard terminology for the head and face. Am J Med Genet A 2009;149A:6–28.
Allanson JE, Biesecker LG, Carey JC, Hennekam RC. Elements of morphology: introduction. Am J Med Genet A 2009;149A:2–5.
Biesecker LG, Aase JM, Clericuzio C, Gurrieri F, Temple IK, Toriello H. Elements of morphology: standard terminology for the hands and feet. Am J Med Genet A 2009;149A:93–127.
Carey JC, Cohen MM Jr, Curry CJ, Devriendt K, Holmes LB, Verloes A. Elements of morphology: standard terminology for the lips, mouth, and oral region. Am J Med Genet A 2009;149A:77–92.
Robinson PN, Mundlos S. The human phenotype ontology. Clin Genet 2010;77:525–534.
Köhler S, Doelken SC, Mungall CJ, et al. The Human Phenotype Ontology project: linking molecular biology and disease through phenotype data. Nucleic Acids Res 2014;42(Database issue):D966–D974.
Robinson PN, Köhler S, Bauer S, Seelow D, Horn D, Mundlos S. The Human Phenotype Ontology: a tool for annotating and analyzing human hereditary disease. Am J Hum Genet 2008;83:610–615.
Girdea M, Dumitriu S, Fiume M, et al. PhenoTips: patient phenotyping software for clinical and research use. Hum Mutat 2013;34:1057–1065.
Richards S, Aziz N, Bale S, et al.; ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405–424.
Rehm HL, Bale SJ, Bayrak-Toydemir P, et al.; Working Group of the American College of Medical Genetics and Genomics Laboratory Quality Assurance Commitee. ACMG clinical laboratory standards for next-generation sequencing. Genet Med 2013;15:733–747.
Gargis AS, Kalman L, Berry MW, et al. Assuring the quality of next-generation sequencing in clinical laboratory practice. Nat Biotechnol 2012;30:1033–1036.
Duzkale H, Shen J, McLaughlin H, et al. A systematic approach to assessing the clinical significance of genetic variants. Clin Genet 2013;84:453–463.
MacArthur DG, Manolio TA, Dimmock DP, et al. Guidelines for investigating causality of sequence variants in human disease. Nature 2014;508:469–476.
Rehm HL, Berg JS, Brooks LD, et al.; ClinGen. ClinGen–the Clinical Genome Resource. N Engl J Med 2015;372:2235–2242.
Jarvik GP, Amendola LM, Berg JS, et al.; eMERGE Act-ROR Committee and CERC Committee; CSER Act-ROR Working Group. Return of genomic results to research participants: the floor, the ceiling, and the choices in between. Am J Hum Genet 2014;94:818–826.
ACMG Board of Directors. ACMG policy statement: updated recommendations regarding analysis and reporting of secondary findings in clinical genome-scale sequencing. Genet Med 2015;17:68–69.
Korf BR, Ledbetter D, Murray MF. Report of the Banbury Summit Meeting on the evolving role of the medical geneticist, February 12-14, 2006. Genet Med 2008;10:502–507.
Kullo IJ, Jarvik GP, Manolio TA, Williams MS, Roden DM. Leveraging the electronic health record to implement genomic medicine. Genet Med 2013;15:270–271.
Manolio TA, Green ED. Leading the way to genomic medicine. Am J Med Genet C Semin Med Genet 2014;166C:1–7.
Burke W, Evans BJ, Jarvik GP. Return of results: ethical and legal distinctions between research and clinical care. Am J Med Genet C Semin Med Genet 2014;166C:105–111.
Jacob HJ, Abrams K, Bick DP, et al. Genomics in clinical practice: lessons from the front lines. Sci Transl Med 2013;5:194cm195.
Vassy JL, Lautenbach DM, McLaughlin HM, et al.; MedSeq Project. The MedSeq Project: a randomized trial of integrating whole genome sequencing into clinical medicine. Trials 2014;15:85.
Acknowledgements
The Clinical Genetics Think Tank was funded by the Centre for Genetic Medicine, Hospital for Sick Children, and the Departments of Pediatrics and Pathology at the Children’s Hospital of Philadelphia, with additional funding from the National Human Genome Research Institute (5UO1HG006546 to I.D.K. and N.B.S.). The authors acknowledge the parent and patient members of the CGTT, M. Hardy, B. Kovalski, H. Kovalski, D. Siciliano, J. Strautnieks, and T. Stoppa, and the support of Joe St. Geme and Bob Doms at the Children’s Hospital of Philadelphia. L.G.B. was supported by the Intramural Research Program of the National Human Genome Research Institute. Funding to cover attendance and accommodation for all participants at both meetings was provided by the Centre for Genetic Medicine, Hospital for Sick Children, Toronto, Canada, and the Departments of Pediatrics and Pathology at the Children’s Hospital of Philadelphia, Philadelphia, Pennsylvania, USA.
Author information
Authors and Affiliations
Corresponding authors
Supplementary information
Supplementary Table S1
(DOC 72 kb)
Rights and permissions
About this article
Cite this article
Bowdin, S., Gilbert, A., Bedoukian, E. et al. Recommendations for the integration of genomics into clinical practice. Genet Med 18, 1075–1084 (2016). https://doi.org/10.1038/gim.2016.17
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gim.2016.17
Keywords
This article is cited by
-
Sibling variation in polygenic traits and DNA recombination mapping with UK Biobank and IVF family data
Scientific Reports (2023)
-
‘Diagnostic shock’: the impact of results from ultrarapid genomic sequencing of critically unwell children on aspects of family functioning
European Journal of Human Genetics (2022)
-
Genetic architecture of complex traits and disease risk predictors
Scientific Reports (2020)
-
Sibling validation of polygenic risk scores and complex trait prediction
Scientific Reports (2020)
-
From Genetic Testing to Precision Medicine in Epilepsy
Neurotherapeutics (2020)