Abstract
Tamoxifen, a widely prescribed drug for the treatment and prevention of breast cancer, is metabolized to more potent metabolites by the cytochrome P450 2D6 (CYP2D6) enzyme. Variants in the CYP2D6 gene can cause patients to be either intermediate or poor metabolizers, thereby rendering tamoxifen treatment less effective. Testing for CYP2D6 gene variants is available in Clinical Laboratory Improvement Amendments–certified clinical laboratories; however, the biological complexity of the variants makes result interpretation and phenotype prediction challenging. This article describes the clinical significance of variants as well as important analytical, interpretative, and reporting issues. It is designed to be a guideline for clinical laboratory professionals in performing tests and interpreting results with respect to CYP2D6 genetic variants.
Genet Med 2012:14(12):990–1000
Similar content being viewed by others
Introduction
Laboratories are beginning to offer pharmacogenetics assays in support of continuing efforts toward more personalized medicine. In patients with breast cancer for whom treatment with tamoxifen is being considered, the clinical validity of CYP2D6 genotyping is not well established. Nonetheless, the results may be helpful in identifying patients who will not respond well to the treatment. This article is intended to help clinical laboratory personnel who are offering or intending to offer CYP2D6 genotyping in patients for whom tamoxifen therapy is being considered. The compound tamoxifen belongs to the class of drugs known as selective estrogen receptor modulators. Our focus here is on laboratory implementation including analytical and clinical validity, available platforms, challenges in the interpretation and reporting of complex test results, and the potential for clinical utility. Clinical CYP2D6 genotyping can also be ordered in the context of other drugs and potential adverse outcomes (e.g., antidepressants, antipsychotics, and codeine). However, a detailed description of implementing CYP2D6 genotyping for these other medications falls outside the scope of this article.
Mechanism of Action and Metabolism of Tamoxifen
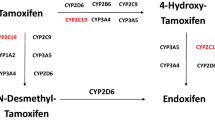
Tamoxifen is one of the most widely used drugs in the treatment and prevention of estrogen receptor–positive breast cancer. It is a chemically nonsteroidal agent, the trans-isomer of a triphenylethylene derivative. The pharmacology of tamoxifen is thought to be linked to the interaction of the parent drug and its metabolites with the estrogen receptor in both breast and nonbreast tissues. This interaction induces a specific conformational change in the estrogen receptor, leading to alteration of downstream signaling pathways, and resulting ultimately in transcriptional and posttranscriptional changes in estrogen-regulated genes, with consequent inhibition of estrogen effects.1 Although the metabolism of tamoxifen in vivo involves several CYP isoforms, cytochrome P450 (CYP)2D6 predominantly catalyzes the formation of the two most potent metabolites of tamoxifen: endoxifen and 4-OH tamoxifen ( Figure 1 ).2 The metabolic activity of CYP2D6 is known to be influenced by the individual’s genotype and by environmental factors (e.g., the ingestion of CYD2D6 inhibitors, which are commonly prescribed). Commonly occurring CYP2D6 genetic variants that lead to low or absent CYP2D6 activity and/or the direct inhibition of CYP2D6 enzyme activity can significantly contribute to interindividual variability in the concentration levels of tamoxifen metabolites.3,4 It has been demonstrated that, despite their contributory roles, other CYP isoforms appear to be less important in tamoxifen metabolism.2
Tamoxifen metabolism. The parent drug tamoxifen (TAM) is converted to the more potent metabolite endoxifen by a complex metabolic route that is ultimately dependent on cytochrome P450 2D6 activity. Source: ref. 4. By permission of Oxford University Press.
CYP2D6 Genotypes
The CYP2D6 gene is located at chromosome 22q13.1. No disease is known to be directly associated with CYP2D6; however, when exposed to an exogenous substance, adverse events may occur. Alleles for the CYP2D6 gene are defined by combinations of variants found on a chromosome and designated using a star (“*”) nomenclature.5 Subsequent variant alleles are numbered in the order in which they are identified and characterized. Within each “*” allele designation, additional variations linked in cis with the defining single-nucleotide polymorphism create subfamilies that are designated alphabetically in the order in which they are described (e.g., *2A, *2B, and *2C). Unlike many heritable disease mutations, each CYP2D6 allele may include several single-nucleotide polymorphisms — a haplotype, rather than a single-site mutation. The *1 (wild-type) allele encodes a fully functioning enzyme and is the most commonly occurring allele in most populations. Benign allelic variants such as *2 and *35 have normal activity, whereas deleterious variants result in null alleles (no protein expressed or nonfunctional) or proteins with altered enzymatic activity. Examples of nonfunctional alleles are *5 (gene deletion) and *4 (splice-site variant). The *10 and *41 variant alleles have partial activity and are referred to as decreased-function or reduced-function alleles. A gene duplication of a functional allele results in increased expression of the active enzyme. Duplication of nonfunctional and reduced-function alleles has also been reported; however, the exact effect of some of these alleles on enzyme activity is difficult to determine.5 A list of variants commonly tested for, with their predicted activities, is shown in Table 1 . It is important to note that all CYP2D6 sequence variants in this article are defined using their common names. They refer to nucleotide positions, with nucleotide 1 being the A of the ATG-translation initiation codon of GenBank Accession Number M33388 (for more information see http://www.cypalleles.ki.se/cyp2d6.htm). Supplementary Table S1 online describes the common names and the corresponding Human Genome Variant Society nomenclature for the major CYP2D6 alleles.
Predicted Phenotype
Genotypes are used to predict phenotypes of poor metabolizer (PM), intermediate metabolizer (IM), extensive (normal) metabolizer (EM), and ultra-rapid metabolizer (UM). Phenotype prediction is dependent on the biallelic expression of alleles: two nonfunctional alleles result in a PM phenotype, whereas two functional alleles, or one functional allele with one decreased-function allele, result in an EM phenotype. IM phenotypes are more difficult to assess, given the lack of consensus in this regard. A nonfunctional allele in combination with a decreased-function allele, or two decreased-function alleles, have some residual enzyme activity and are considered to be IM phenotypes. A nonfunctional allele in combination with a functional allele may be considered to be an EM phenotype, consistent with an autosomal recessive inheritance. However, such genotypes would confer only 50% activity as compared to two copies of functional alleles, and could also be considered as IM phenotypes.6 Another term used for this genotype is “heterozygous extensive”7; this nomenclature may further confuse the issue. The term “heterozygous” simply implies that there are two different alleles present but does not imply any difference in activity, and could therefore be misleading. CYP2D6 gene duplications also result in several possible phenotype scenarios. Duplication of functional alleles results in the UM phenotype. However, duplication of nonfunctional alleles (e.g., *4) has also been reported. Not all platforms identify whether a heterozygous duplicated allele is nonfunctional, and therefore the results of duplication with variants would need to be interpreted as “indeterminate” unless familial studies are performed for haplotype analysis ( Figure 2 ). Also, there is currently no commercial assay that can detect the number of duplicated alleles when duplication is present. This too, can affect the phenotype, because multiduplication (more than two copies) of a functional allele can lead to ultra-rapid metabolism regardless of the other allele. Laboratories must consider these complexities, the capability of the assay, and the current state of knowledge, and should provide a clear explanation in the genotype test report about how a given phenotype prediction has been arrived at.
Pedigree showing possible allele patterns for *4 and *10 alleles in parents and child. In the pattern depicted, the child inherits the *4 allele, which is duplicated, from his mother and the *10 allele from his father. The resulting *4/*10 genotype would generate calls for each nucleotide polymorphism as follows: 100C>T (T/T), 1661G>C (C/C), 2850C>T (T/T), and 4180G>C (C/C), and heterozygous for 1846G>A (G/A), plus a duplication.
There is only limited accuracy in calling an allele normal/functional on the basis of the nucleotide sequence detected. This is because genotyping is not comprehensive enough to identify all currently known sequence variations. Rather, genotyping identifies only the variants for which the assay is specifically designed. For example, sequence variants located in the intronic or regulatory regions, which are not ascertained in the assay, may also affect gene expression. CYP2D6 regulatory elements, and genomic variants that alter their functions, are not yet well understood. The *2 allele is detected by all commercial platforms and is considered to be functional. The *2A subtype is characterized by the additional promoter variant −1584G>A, which may affect expression. Increased expression of this allele has been associated with some substrates, but its effect on tamoxifen metabolism is not well understood. Gene conversion—a mechanism of mutation transfer between the pseudogene and the gene—may also fail to be analyzed by a given platform. Gene conversions may be difficult to detect and this may be a limitation of particular platforms. For example, the gene-converted *36 nonfunctional allele may mistakenly be called a *10 decreased-function allele.5
Critical to phenotype interpretation is a good understanding of the functional consequences of a given variant as regards enzyme activity. More specifically, how does the sequence variant affect the behavior of the enzyme? The National Academy of Clinical Biochemistry (NACB) Laboratory Medicine Practice Guidelines for Pharmacogenetics recommend that pharmacogenetics assays should include testing for genetic variants with either (i) a well-defined influence on enzyme function or (ii) a clear relationship between the variant and an observable influence on drug pharmacokinetics, pharmacodynamics, or toxicology.8 The rationale is that, when there is a clear, mechanistic, cause–effect relationship between the variant and the end point (e.g., metabolic clearance rate), the phenotype can be predicted with greater reliability. The functional consequences of sequence changes are varied. A single variant can abolish protein activity by itself (such as the 1846G>A variant of *4) or may do so in combination with another single variant. Alternatively, a variant such as 100C>T may decrease protein activity without abolishing it, leading to “decreased function” alleles, namely *10. Also, even when a sequence change by itself has no effect on the protein, it may be in linkage disequilibrium with others (as part of a haplotype), thereby affecting protein activity. The ability to identify the variant that is causal to the functional end point is essential to assay design as well as to assignment of the most appropriate phenotype.
Other challenges in predicting phenotype from genotype test results involve substrate or drug specificity. Sequence variants can change substrate specificity rather than enzymatic activity; therefore the metabolism of one drug cannot be assumed to be identical to that of another.9 Some allelic variants (e.g., CYP2D6*17) are associated with substrate-dependent decreases in catalytic properties.10,11,12,13 Variant proteins may decrease the incorporation of heme into, or the stability of, the CYP2D6 apoprotein (e.g., when conserved elements common to the heme binding site or the planar pocket of CYP are affected).
Differences may also exist between in vitro and in vivo measurements. If drugs are also metabolized by other enzymes, in vitro experiments may not reflect the real activity of a certain allele. For example, the variant 2064G>A (new nomenclature: 1976G>A) has decreased activity toward debrisoquine in vivo but absent activity with bufuralol in vitro.14,15 However, its effect on tamoxifen metabolism remains undetermined.
Clinical Validity and Clinical Utility
Currently, in both premenopausal and postmenopausal patients with invasive breast cancer involving estrogen receptor–positive tumors, long-term therapy with tamoxifen is the standard treatment.16 This treatment reduces the recurrence risk by ~50%.17 The clinical outcome of interest is recurrence of cancer, but an intermediate end point could be endoxifen levels. Several studies have shown that circulating levels of endoxifen are lower in the presence of fewer functional CYP2D6 alleles.4,18 If lower endoxifen levels are associated with reduced effectiveness of tamoxifen therapy, CYP2D6 genotyping may provide useful information in personalizing cancer treatments. It has been suggested that patients with no functional CYP2D6 alleles should be offered alternatives to tamoxifen.19
Circulating plasma levels of endoxifen are considered a surrogate outcome for clinical validity, but an analysis of these levels stratified by metabolic phenotype could provide insight into whether higher levels actually do translate into improved survival. We extrapolated data from figures and summarized the statistics from two published reports.4,19 In both studies, the women were given 20 mg/q.d. of tamoxifen, and none were reported as receiving CYP2D6 inhibitors. For this literature review, PMs were defined in terms of the presence of two nonfunctional alleles. Although no consensus exists in this regard, for the purposes of this review we defined IMs as those having two decreased-function alleles, one decreased-function allele and one nonfunctional allele, or one nonfunctional allele and one functional allele. EMs were defined as those having two functional alleles or one functional allele and one decreased-function allele. Less well-defined or infrequent allele combinations were omitted from the analysis (e.g., UMs). The reported mean values and standard deviations were similar in the two reports, and the individual observations were combined and fitted to a Gaussian distribution after a logarithmic transformation. Figure 3 shows the overlapping distributions of plasma endoxifen (x-axis) in the PM, IM, and EM groups, both in relative and absolute terms. In Figure 3a , the distribution for each of the three metabolic groups has the same area, so that shape and position can be directly compared. Figure 3b may be more relevant to the clinical scenario, in that the prevalence of each group is taken into account, with the areas of the distribution curves for the PM, IM, and EM groups being in the ratio of 1:4:9 (~7, 29, and 64%, respectively). Of note, many of the lower values occurred among patients in the lower tails of the IM and EM groups, overlapping with the PM group. Possible explanations for this finding are unreported CYP2D6 inhibitors, other CYP2D6 variants not detected by the assays used, and variants with large effect sizes in other genes involved in tamoxifen metabolism.
A graphic representation of the distribution of plasma endoxifen levels in women treated with tamoxifen, stratified by the three main cytochrome P450 2D6 metabolic phenotypes. The data sources and analytic methods are described in the text. (a) The relative distributions (same area under the curve for each distribution) for the poor (PM), intermediate (IM), and extensive (EM) metabolizer phenotypes. Although the mean levels are clearly different, there is important overlap, especially between the IM and EM phenotypes. (b) The same data, but incorporating the prevalence of each of the phenotypes into the figure (area under the distribution represents the proportion of the population with that phenotype).
Currently, there is no established “lower limit” of circulating endoxifen below which the treatment would be ineffective. Figure 3b can be used to help evaluate whether genotyping might be a useful surrogate for a nontherapeutic value of circulating endoxifen; for example, if a hypothetical level of <20 nmol/l is considered nontherapeutic, CYP2D6 genotyping would be a screening test to identify patients who are predicted to have nontherapeutic endoxifen levels. The outcome is determined by the endoxifen level (e.g., <20 and ≥20 nmol/l), while the screening test is CYP2D6 genotype–inferred metabolic phenotype (PM = positive; IM and EM = negative). Using the same data as for Figure 3 , Table 2 shows the detection rate (proportion of all patients having nontherapeutic levels detected using PM as a positive test) along with the false-positive rate (proportion of patients with PM genotype having therapeutic levels). Therefore, using 20 nmol/l as the definition of a therapeutic level, identifying the PM genotype by CYP2D6 genotyping has a detection rate of ~69%, and a corresponding false-positive rate of between 3 and 4%. That is, ~31% of the patients with nontherapeutic levels of endoxifen would not be identified, and 3–4% of those classified as nontherapeutic would actually lie within the therapeutic range. Drug models show that, ultimately, only small levels of endoxifen are needed to block all receptor sites,17 implying that the suggested cause–effect relationship between CYP2D6 status and sub-therapeutic endoxifen concentrations may be only part of the story.
Several studies have examined the relationship between disease recurrence and CYP2D6 genotype, with widely varying results. Some have shown a strong positive association, whereas others have found none; some have even found a negative association with EMs. In postmenopausal patients taking adjuvant tamoxifen, several studies have shown that impaired CYP2D6 metabolism, caused either by genetic variation12,20,21,22 or by enzyme inhibition,11 is a predictor of worse outcomes in breast cancer, including statistically significant shorter time to recurrence and poorer relapse-free survival. However, overall survival was not found to be significantly different among the various groups. Some subsequent studies of the association between CYP2D6 status and breast cancer outcomes in tamoxifen-treated patients in preventative12 and metastatic23 settings have supported the consistency of these data. However, other adjuvant studies have refuted such data with evidence that tamoxifen-treated women with CYP2D6 variants have similar outcomes24 or even better outcomes13,25 than women with normal CYP2D6 genotypes. Investigators in the Consortium on Breast Cancer Pharmacogenetics have recently suggested that these studies, in addition to being confounded by variations in treatment strategies and settings, were all small and predominantly retrospective in nature, resulting in a low confidence level.26 Given this uncertainty, the Consortium on Breast Cancer Pharmacogenetics investigators recommend against routine CYP2D6 genotyping for patients who are candidates for tamoxifen, while recognizing that circumstances may exist where such knowledge could be helpful. They do recommend, however, that patients treated with tamoxifen avoid comedications that are known to inhibit CYP2D6 metabolism.26 Meanwhile, a recent study in >1,300 patients showed significant disease-free and event-free survival in patients with early-stage breast cancer treated with tamoxifen, and that those with genotype-predicted EM phenotypes had significantly lower recurrence rates and better disease-free and event-free survival than those who were predicted as being IM or PM phenotypes. As with other studies, overall survival was not significantly different among the groups.27
It is difficult to establish clear evidence of the clinical validity of CYP2D6 genotyping in patients for whom tamoxifen treatment is being considered. Studies designed to settle the issue may or may not be feasible. If one overarching study is not possible, several studies may be required to achieve a supportive level of evidence.17 There is potential for CYP2D6 genotyping to have clinical utility, but the current evidence base is heterogeneous and based on relatively small sample sizes. The 2009 National Comprehensive Cancer Network breast cancer guidelines do not include any recommendations concerning treatment modifications based on the outcome of CYP2D6 genotyping.16
Although clinical validity in terms of survival has not been established, the strong association of CYP2D6 genotype with endoxifen levels suggests that genotyping is a practical approach to predicting tamoxifen metabolizer phenotypes.4,18 As commercial products become available, clinical laboratories are implementing CYP2D6 genotyping and offering it as a clinical test for tamoxifen as well as for other CYP2D6-metabolized medications.
Commercial Platforms
Several commercial CYP2D6 genotyping assay platforms are available ( Table 3 ). Although all of them test for the presence of most common variants, they differ with respect to the range of variants detected. They also differ in how they call alleles and in whether or not an allele designation is provided as part of the result. Also, for some combinations of alleles, the classification into metabolic phenotypes is not yet standardized. Here we describe and compare three commercial platforms ( Table 1 ).
AmpliChip CYP450 Test—background and variants tested
The AmpliChip CYP450 Test (manufactured by Roche Molecular Systems, Branchburg, NJ; distributed by Roche Diagnostics, Indianapolis, IN) is an oligonucleotide microarray hybridization method of identifying an individual’s CYP2D6 and CYP2C19 genotypes from genomic DNA extracted from a whole-blood sample. (Note: CYP2C19 has not been noted to be an important pathway in tamoxifen metabolism, and its use in this context will not be specifically addressed). This assay was cleared for clinical use by the US Food and Drug Administration (FDA) at the beginning of 2006.28
The AmpliChip CYP450 Test detects the most common and phenotypically relevant CYP2D6 and CYP2C19 variants observed across various ethnic groups. Twenty CYP2D6 alleles (*1(presumed),*2ABD,*3,*4ABDJK,*5,*6ABC,*7,*8,*9,*10AB,*11,*15,*17,*19,*20,*29,*35,*36,*40, and *41), seven CYP2D6 duplications (*1XN,*2XN,*4XN,*10XN,*17XN,*35XN, and *41XN), and three CYP2C19 alleles (*1 (presumed),*2, and *3) are analyzed and reported. The microarray utilizes approximately 240 unique probes to accurately detect each variant with a high level of redundancy (>15,000 different oligonucleotide probes). The probes are synthesized on a glass substrate to analyze both sense and antisense strands of amplified target DNA.
Description of the assay
The AmpliChip CYP450 Test amplifies the CYP2D6 and CYP2C19 genes in two separate reactions that are subsequently pooled after PCR amplification. The reaction containing CYP450 Primer Mix A uses primers that generate amplified product encompassing the promoter region and coding regions of the CYP2D6 gene, and a CYP2D6 gene duplication-specific product, when present, in the specimen or control. The reaction containing CYP450 Primer Mix B uses primers that generate amplified product encompassing exon 4 and exon 5 of the CYP2C19 gene, and a CYP2D6 gene deletion-specific product, when present, in the specimen or control. The pooled DNA amplicons from the two independent amplification reactions are fragmented by DNAse I and alkaline phosphatase; this is done in order to generate small, single-stranded DNA fragments with an average size of 50–200 nucleotides, and to destroy the residual deoxyribonucleotide triphosphates from the amplification reactions, respectively. The fragmented DNA amplicons are subsequently labeled with biotin at their 3′ termini by terminal transferase, using AmpliChip TdT Labeling Reagent as substrate. The biotin-labeled CYP450 target DNA fragments are hybridized to the oligonucleotides on the AmpliChip CYP450 Microarray, using the Affymetrix GeneChip Fluidics Station 450Dx (Santa Clara, CA) and an AmpliChip CYP450-specific protocol. The hybridized AmpliChip CYP450 Microarray is washed and stained with a streptavidin-conjugated fluorescent dye (phycoerythrin). After staining, the AmpliChip CYP450 Microarray is scanned by an Affymetrix GeneChip Scanner 3000Dx using a laser that excites the fluorescent label that is bound to the hybridized CYP450 target DNA fragments. The amount of emitted light is proportional to the amount of bound target DNA at each location on the probe microarray.
Data analysis
Data analysis is performed using the Affymetrix GeneChip Operating Software (GCOSDx Version 1.1.3; Santa Clara, CA) and the AmpliChip CYP450_US Data Analysis Software v2.1, or by means of the new operating system, Affymetrix Molecular Diagnostics Software for the Affymetrix GeneChip System 3000Dx v.2 and the AmpliChip CYP450 Data Analysis Software v3.1. The presence or absence of the particular polymorphisms listed in Table 1 determines the CYP2D6 and CYP2C19 allele and predicts the enzymatic activity of the gene product. The report generated by the CYP450 Data Analysis Software v2.1 for each particular sample includes the genotype call for the CYP2D6 and CYP2C19 genes and the predicted phenotypes. The upgraded version of the software (CYP450 Data Analysis Software v3.1) allows independent reporting of either the CYP2D6 or CYP2C19 genotypes and phenotypes, or both. The AmpliChip CYP450 Test classifies individuals into one of four CYP2D6 phenotypes: UM, EM, IM, and PM, and two CYP2C19 phenotypes: EM and PM. The software also provides a table showing the interrogated alleles, and a site and call list that describes the polymorphism site identified by the nucleotide position and base change as well as the polymorphism call.
Advantages of the platform
As compared to other methods, the Amplichip CYP450 Test allows for multiple simultaneous testing of many alleles, and potentially reduces technician time by automating the process. It utilizes ~240 probes to detect each polymorphism with wild-type and mutant probe sets, thereby reducing the possibility of missing a nontargeted mutation and resulting in greater confidence in the *1 allele call. Another advantage is the availability of software that analyzes the data using specific algorithms for genotype information and predicted phenotypes. Finally, the assay can be performed with input DNA levels as low as 25 ng for CYP2D6 and 2.5 ng for CYP2C19.
Luminex—background and variants tested
Luminex is a bead array platform based on allele-specific primer extension (ASPE) and hybridization to oligonucleotide-bound microspheres. The current version of the assay (Version 3) interrogates 22 variants (including 20 nucleotide positions, as well as gene deletion and duplication) corresponding to 17 variant alleles. The variants that can be detected using this array are *1 (presumed), *2, *3, *4, *5, *6, *7, *8, *9, *10, *11, *12, *14, *15, *17, *29, *35, and *41. This assay was cleared for clinical use by the FDA in August 2010.29
Description of the assay
The xTAG CYP2D6 assay (Luminex, Austin, TX) consists of two separate multiplex PCR reactions with amplicons >1 kilobase in size. The large amplicons are necessary for avoiding co-amplification of pseudogenes that could interfere with the assay. After PCR, the products are treated with exonuclease I and shrimp alkaline phosphatase to remove unincorporated deoxyribonucleotide triphosphates and primers, respectively. The next step is an ASPE reaction. Each position to be interrogated has two primers associated with it, designed to have the 3′ base specific to either the wild-type or the variant allele. Each allele-specific primer also has a 5′ tag sequence necessary for hybridization to the bead array. Allele-specific primer extension and subsequent incorporation of biotin-deoxyribose cytosine triphosphate occurs only from a perfectly matched 3′ end of the allele-specific primer. The next step is hybridization of the ASPE products to the microspheres. Each bead of the array is composed of red and infrared fluorophores, and is tagged with a universal anti-tag sequence complementary to the allele-specific primer tags. The bead-ASPE hybridization products are incubated with an R-phycoerythrin-strepavidin conjugate reporter solution, and fluorescence intensities are measured using the Luminex xMAP IS system. Two lasers are used to detect the unique mixtures of fluorescent signals that identify each microsphere and any phycoerythrin signal that may have been produced.29
Data analysis
The xTAG data analysis software provides a wild-type (WT), heterozygous (HET), or homozygous variant (MUT) call at each variant position. The software indicates the variants with which the identified alleles are commonly associated; however, the information is not all encompassing, given that it assigns neither haplotypes nor predicted phenotypes. It is the user who must assign the genotype on the basis of the software-generated calls at each variant position. Although the assay can detect the presence of a duplication, it cannot detect how many copies of the gene are present, and cannot predict which allele is duplicated in a heterozygous sample. This is of particular importance for phenotype designation in situations wherein the heterozygous alleles include a nonfunctional or decreased-function allele. For example, in a patient carrying the CYP2D6*1/*4 genotype with gene duplication, either the *1 or the *4 allele may be the duplicated allele. If the *1 allele is duplicated (*1XN/*4), the patient would have two functional copies of the gene (assuming N = 2) and would thereby be predicted to be an EM phenotype. However, if the *4 is the duplicated allele (*1/*4XN), it would continue to remain nonfunctional, and the patient would have only one functional allele; this would result in an IM-to-EM phenotype.
Advantages of the platform
The Luminex assay is performed in a 96-well format for PCR, and the plate is transferred directly to the plate reader. Signal detection is very rapid, with all 96 samples being completed in ~10 min. The platform is well suited for high throughput.
AutoGenomics—background and variants tested
The AutoGenomics (Carlsbad, CA) platform is a film-based microarray tested on the INFINITI Analyzer.30 The analyzer includes a confocal microscope with two lasers that detect the amount of fluorescently labeled target that is bound to each probe on the microarray. Two separate CYP2D6 assays are currently available from AutoGenomics. The CYP2D6T assay includes detection of the *2, *3, *4, *5, *6, *7,*8, *9, *14, *29, and *41 variants. The CYP2D6I assay includes detection of the *2, *3, *4, *5, *6, *7, *8, *9, *10, *12, *14, *17, *29, and *41 variants, as well as of duplication alleles.
Description of the assay
DNA is first PCR-amplified with primers specific to the CYP2D6 locus, designed to avoid amplification of the pseudogenes. After PCR, the products are treated with exonuclease I/shrimp alkaline phosphatase to remove excess primer. The treated products are then loaded onto the INFINITI Analyzer. ASPE allows for labeling and signal amplification, and the fluorescently labeled ASPE products are hybridized to the BioFilmChip microarray. The labeled products bind specifically to their complementary microarray locations. The microarray is scanned by the internal confocal microscope
Data analysis
The fluorescence levels detected by the microscope are converted into genotype data by the Qmatic software (AutoGenomics, Carlsbad, CA). As with similar ASPE-based methods, the specific allele that is duplicated cannot be determined in a heterozygous sample. Allelic ratios are determined, and the software calls homozygous mutation (M), wild-type (W), or heterozygous (H) for each allele.
Advantages of the platform
The platform includes liquid handling, and therefore all post-PCR steps are automated. The platform is suited for low-to-moderate throughput, with a capacity to analyze 24 samples at a time.
Analysis
Analyses for multiple variants pose complexities, and there are several challenging issues with respect to all these platforms. Some of the complexities that may affect interpretation are discussed here.
Although the entire industry uses “haplotype/star nomenclature” when calling CYP2D6 genotyping results, this standard practice is misleading because haplotyping assays, in contrast to genotyping assays, are intended to look at a “larger picture” of haplotypic block segregation. The assays described in the CYP2D6 genotypes section are genotyping assays, and yet alleles are often defined in terms of the combination of variants in the haplotype ( Table 1 ). Although molecular haplotyping techniques have been described, they are complex and rarely used in clinical molecular laboratories. Instead, variants are detected individually and then, not always correctly, combined for a presumed haplotype and allele on the basis of published data. For example, the nonfunctional *4 allele is defined by several variants, including the 100C>T and 1846G>A. The 1846G>T variant, characteristic for the *4 allele, causes a splice-site change; this change results in a frameshift that destroys the activity of the enzyme. The 100C>T variant may also be present without the accompanying 1846G>A change. This is a milder variant, resulting in the *10 decreased-function allele. Although 1846G>A can exist on a chromosome without 100C>T (*4M), this allele is rare. When both variants are seen as heterozygotes, the probable genotype is *4 heterozygous (one copy of a nonfunctional allele) given that 1846G>A and 100C>T are presumed to be on the same chromosome. Some instruments may call each DNA variant separately, without combining variant combinations into a haplotype. Without haplotyping, one copy each of 100C>T and 1846G>A could be misinterpreted as being *4/*10 compound heterozygous (one copy of a nonfunctional allele and one copy of a decreased-function allele). A presumed *4/*10 combination would appear as two copies of 100C>T (homozygous T/T) with only the polymorphic nucleotide T called and not C, and with one copy of 1846G>A (heterozygous GA). In addition, *2 is defined by 2850C>T and 4180G>C, and is associated with an EM phenotype. The defining variant in *41 is 2988G>A with 2850C>T and 4180G>C in the haplotype, and is associated with decreased enzymatic activity. When all three variants are present in the heterozygous state, the correct star allele genotype is a *41 heterozygote. If 2988G>A is heterozygous G/A, 2850C>T is homozygous T/T, and 4180G>C is homozygous C/C, the correct star allele genotype is a *2/*41 compound heterozygote. When using platforms that identify a star allele for each nucleotide change, laboratories will need to assign the correct haplotypes for the allele assignments ( Table 1 ).
Occasionally, combinations of variants are detected that are not consistent with published alleles. In addition, if a duplication is detected with a nonfunctional or decreased-function allele, some of the assays will be unable to determine which of the alleles (the functional allele or the nonfunctional/decreased-function allele) is duplicated. In both cases, family studies may help determine the haplotype (chromosome phase). Figure 2 gives an example of a family study and shows the allelic patterns for a *4/*10 duplication in the proband and each parent.
As with many genetic tests, not all variants are detected by assays developed (commercially or by laboratories) for targeted mutation panels. In the case of rare alleles that the assay is not designed to detect, the software of the assay may default to an allele that is most genetically similar, or call as “no variant detected.” For example, a platform may not be designed to detect the allele defining variant 2573insC that is present in the CYP2D6*21B allele, predicted to be a loss-of-function allele. In specimens containing this rare insertion allele, it may be identified as the closely related CYP2D6*2 allele, a normal-function allele. Another example: the *41 allele has been recently described as a decreased-function allele defined by the 2988G>A variant. Assays that do not detect this variant will make a call based on the combined presence of other variants in the haplotype, namely, 1584C, 1661G>C, 2850C>T, and 4180G>C, which are also found in *2 subtypes.
Although CYP2D6 variants are found in all ethnic groups, some alleles are seen more frequently in specific populations. For example, *3 is more prevalent in Caucasian populations than in Asian or African populations, whereas *10 is more prevalent in Asian populations. Table 4 summarizes allele frequencies of the more common variants in Caucasian, Asian, and African-American populations.
Reporting
Because the reason for testing is to predict the metabolic phenotype on the basis of the presence or absence of the nucleotide variation, laboratory reports should give the predicted phenotype while reporting the result, along with details of the alleles detected. When none of the targeted variants is detected, this is consistent with the *1 or normal allele. However, because not all possible variants are included in the testing panel, rare variants that are present may go undetected. For this reason, the result is more accurately phrased as “none of the targeted variants detected.” However, laboratory reports often use the terminology “Negative” or “*1” to indicate this situation. The Amplichip platform that was cleared by the FDA for commercial use interprets “no variant allele is detected” as a *1 or normal allele. It may therefore be required that this nomenclature be used when seeking FDA clearance for an assay.
Although the use of standard nomenclature is ideal, common names (such as the star nomenclature) may be more readily recognized. For example, standard nomenclature of the *4 allele is based on the presence of the 1846G>A defining variant, although other haplotypic variants could also be present. A *4/*41 genotype, when expressed in standard nomenclature, may appear as (depending on the platform used):
CYP2D6: 100C>T, 1661G>C, 1846G>A, 4180G>C/1661G>C, 2850C>T, 2988G>A, 4180G>C.
Using only allele-defining variants, the simplest form in standard nomenclature would appear as CYP2D6: 1846G>A/2988G>A.
Because laboratories use different platforms and may detect different variants, the report should include all the variants that have been tested for, the methodology, the details of the assay, and its limitations. The NACB’s Laboratory Medicine Practice Guidelines8 are in agreement in recommending that the list of nucleotide variants tested and detected be included in the report in the event that nomenclature systems change over time. The College of American Pathologists Molecular Pathology laboratory checklist also requires the reporting of all gene loci tested (MOL36000;31).
CYP2D6 reports should have an interpretative component, which may include additional recommendations such as use of alternative therapies, altered dosage, avoidance of known CYP2D6 inhibitors, and other confounding clinical factors, if known. The CAP Molecular Pathology checklist requires that a test report include not only the variants tested for and detected, but also the analytic and clinical interpretation (if appropriate) of the results (MOL.36000). The interpretative report should effectively convey the result and its clinical ramifications to a “nonexpert physician.”31 NACB guidelines concur that the information provided in the interpretative report should be “useful for guiding therapeutic management and decision making.”8 The inclusion of suggestions for patient-specific dosages is not encouraged in reports because laboratories typically do not have relevant information (such as the patient’s height and weight and concomitant medications) that can affect dosing. However, the reports could indicate whether lower or higher doses, or alternative drugs, may be considered.
The NACB also suggests that, in order to provide optimal interpretative guidance, the report should include, when possible, specific information relating to the medications involved in the clinical situation. This implies that when a laboratory knows that CYP2D6 genotyping has been ordered for tamoxifen therapy, interpretative information specific to tamoxifen should be provided. The rationale for this recommendation is that the accuracy of phenotypic prediction is dependent on the variants detected as well as on the drug substrate in question. In addition, drug–gene and drug–enzyme interactions can dramatically alter the phenotype. For example, a patient on tamoxifen therapy who is also prescribed a strong CYP2D6 inhibitor (e.g., paroxetine) may be at increased risk for tamoxifen treatment failure because of inability to produce adequate levels of endoxifen. Even a patient who would be predicted to be an EM on the basis of genotype could appear as a phenocopy of a PM because of the strong inhibition of CYP2D6 by paroxetine. In this example, the patient’s genotype would not be the best predictor of phenotype.
A “nonexpert physician” may not fully appreciate these confounding issues unless they are included in the genotyping report. Perhaps the most significant limitation to providing such patient-specific information, however, is the fact that laboratories rarely know what drugs are involved in the clinical scenario, and have limited means of gaining such information. An alternative approach could be to consider inclusion of generalized interpretative components in the reports, mentioning how these confounding factors could potentially affect the phenotype status of a patient. Examples of CYP2D6 genotype reports in the recommended format32 are given in Appendix A online.
Reference Materials
Well-characterized reference materials (RMs) are fundamental to laboratory quality assurance programs, internal quality assurance activities such as quality control, test development/validation, and external assessment by proficiency testing (PT). The selection of appropriate material is based on the needs of the assay, test methodology, and availability. To improve the public availability of characterized RMs, the Centers for Disease Control and Prevention, in partnership with the clinical testing community, has established the Genetic Testing Reference Materials Coordination Program.33 Its goal is to improve the supply of publicly available and well-characterized genomic DNA that can be used as reference materials for PT, quality control test development/validation, and research studies. In conjunction with the Association for Molecular Pathology and the College of American Pathologists (CAP), the Genetic Testing Reference Materials Coordination Program coordinated the characterization of 107 DNA samples for clinically relevant polymorphisms for pharmacogenetic loci including CYP2D6.34 Other loci that have been characterized include CYP450 (2C9, 2C19, 3A4, and 3A5), VKORC1, UGT1A1, MTHFR, NAT2, EPXH1, ABCB1, HLA B, and KIF6. These DNA samples are available from the Coriell Cell Repositories (Camden, NJ).
Several other types of RMs for CYP2D6 testing are available for public use, in addition to the cell lines and genomic DNAs described here. Currently, there are FDA-cleared products that have the following characterized alleles: *1, *2M, *3A, *4A, *5, *6B, *17, *29, *41, and *2AxN.35 There are also Research Use Only products that have the following characterized alleles: *1 (presumed), *2A, *4, *4A, *5, *6B, *7, *9, *10B, *17, *35, *41, *1XN, and *4XN. The RMs have been validated by bidirectional sequencing.35
Proficiency Testing
For quality control and assurance purposes, and as part of CLIA certification, laboratories participate in proficiency testing. A pharmacogenetics module that includes CYP2D6 is administered by the CAP as part of a joint program with the American College of Medical Genetics and Genomics (ACMG). The challenge assesses both analytical and interpretative aspects of testing of three samples, twice a year. Although this challenge is not yet graded, the responses are summarized, along with a discussion of the intended responses to help educate participants.36
Conclusions
Laboratories should understand the challenges involved in establishing the clinical validity of CYP2D6 testing for patients for whom tamoxifen therapy is being considered. Laboratories that are planning to offer CYP2D6 testing should give adequate consideration regarding the platform to be used and the alleles to be tested for. Although a variety of platforms are available to fit individual laboratory workflows, each has technical challenges beyond those that are typical for molecular genetics assays, and it is essential that a laboratory that intends to offer the test must understand these challenges. Interpretation is not straightforward, and laboratories should be familiar with allele nomenclature and haplotypes to properly interpret the results. The assay must be validated and controlled for with available reference material, and routinely checked through proficiency testing. Reports should accurately reflect the genotypes as well as the predicted phenotypes so as to help clinicians understand the clinical ramifications of the results, but always within the context of the known limitations of the assay.
Disclosure
The following authors direct laboratories that perform clinical testing for CYP2D6 alleles: E.L., V.M.P., K.R., and S.A.S. The other authors declare no conflict of interest.
References
Lau CK, Subramaniam M, Rasmussen K, Spelsberg TC . Rapid induction of the c-jun protooncogene in the avian oviduct by the antiestrogen tamoxifen. Proc Natl Acad Sci USA 1991;88:829–833.
Desta Z, Ward BA, Soukhova NV, Flockhart DA . Comprehensive evaluation of tamoxifen sequential biotransformation by the human cytochrome P450 system in vitro: prominent roles for CYP3A and CYP2D6. J Pharmacol Exp Ther 2004;310:1062–1075.
Stearns V, Johnson MD, Rae JM, et al. Active tamoxifen metabolite plasma concentrations after coadministration of tamoxifen and the selective serotonin reuptake inhibitor paroxetine. J Natl Cancer Inst 2003;95:1758–1764.
Jin Y, Desta Z, Stearns V, et al. CYP2D6 genotype, antidepressant use, and tamoxifen metabolism during adjuvant breast cancer treatment. J Natl Cancer Inst 2005;97:30–39.
Ingelman-Sundberg M, Daly AK, Nebert DW (eds). The Human Cytochrome P450 (CYP) Allele Nomenclature Database. 2009. http://www.cypalleles.ki.se/. Accessed 26 October 2010.
Kirchheiner J, Nickchen K, Bauer M, et al. Pharmacogenetics of antidepressants and antipsychotics: the contribution of allelic variations to the phenotype of drug response. Mol Psychiatry 2004;9:442–473.
Eap CB, Lima CA, Macciardi F, Woggon B, Powell K, Baumann P . Steady state concentrations of the enantiomers of mianserin and desmethylmianserin in poor and in homozygous and heterozygous extensive metabolizers of debrisoquine. Ther Drug Monit 1998;20:7–13.
Valdes RV, Payne D, Linder MW, et al. Guidelines and Recommendations for Laboratory Analysis and Application of Pharmacogenetics to Clinical Practice: DRAFT. National Academy of Clinical Biochemistry Laboratory Medicine Practice Guidelines. http://www.aacc.org/members/nacb/LMPG/OnlineGuide/PublishedGuidelines/LAACP/Documents/PGx_Guidelines.pdf. Accessed 6 October 2010.
Wang B, Yang LP, Zhang XZ, Huang SQ, Bartlam M, Zhou SF . New insights into the structural characteristics and functional relevance of the human cytochrome P450 2D6 enzyme. Drug Metab Rev 2009;41:573–643.
Goetz MP, Rae JM, Suman VJ, et al. Pharmacogenetics of tamoxifen biotransformation is associated with clinical outcomes of efficacy and hot flashes. J Clin Oncol 2005;23:9312–9318.
Goetz MP, Knox SK, Suman VJ, et al. The impact of cytochrome P450 2D6 metabolism in women receiving adjuvant tamoxifen. Breast Cancer Res Treat 2007;101:113–121.
Bonanni B, Macis D, Maisonneuve P, et al. Polymorphism in the CYP2D6 tamoxifen-metabolizing gene influences clinical effect but not hot flashes: data from the Italian Tamoxifen Trial. J Clin Oncol 2006;24:3708–9; author reply 3709.
Wegman P, Elingarami S, Carstensen J, Stål O, Nordenskjöld B, Wingren S . Genetic variants of CYP3A5, CYP2D6, SULT1A1, UGT2B15 and tamoxifen response in postmenopausal patients with breast cancer. Breast Cancer Res 2007;9:R7.
Daly AK, Leathart JB, London SJ, Idle JR . An inactive cytochrome P450 CYP2D6 allele containing a deletion and a base substitution. Hum Genet 1995;95:337–341.
Marez D, Legrand M, Sabbagh N, et al. Polymorphism of the cytochrome P450 CYP2D6 gene in a European population: characterization of 48 mutations and 53 alleles, their frequencies and evolution. Pharmacogenetics 1997;7:193–202.
The National Comprehensive Cancer Network (NCCN). http://www.nccn.org/professionals/physician_gls/PDF/breast.pdf. Accessed 29 October 2010.
Lash TL, Lien EA, Sørensen HT, Hamilton-Dutoit S . Genotype-guided tamoxifen therapy: time to pause for reflection? Lancet Oncol 2009;10:825–833.
Borges S, Desta Z, Li L, et al. Quantitative effect of CYP2D6 genotype and inhibitors on tamoxifen metabolism: implication for optimization of breast cancer treatment. Clin Pharmacol Ther 2006;80:61–74.
Goetz MP, Kamal A, Ames MM . Tamoxifen pharmacogenomics: the role of CYP2D6 as a predictor of drug response. Clin Pharmacol Ther 2008;83:160–166.
Kiyotani K, Mushiroda T, Sasa M, et al. Impact of CYP2D6*10 on recurrence-free survival in breast cancer patients receiving adjuvant tamoxifen therapy. Cancer Sci 2008;99:995–999.
Ingle JN, Twito DI, Schaid DJ, et al. Combination hormonal therapy with tamoxifen plus fluoxymesterone versus tamoxifen alone in postmenopausal women with metastatic breast cancer. An updated analysis. Cancer 1991;67:886–891.
Schroth W, Antoniadou L, Fritz P, et al. Breast cancer treatment outcome with adjuvant tamoxifen relative to patient CYP2D6 and CYP2C19 genotypes. J Clin Oncol 2007;25:5187–5193.
Lim HS, Ju Lee H, Seok Lee K, Sook Lee E, Jang IJ, Ro J . Clinical implications of CYP2D6 genotypes predictive of tamoxifen pharmacokinetics in metastatic breast cancer. J Clin Oncol 2007;25:3837–3845.
Nowell SA, Ahn J, Rae JM, et al. Association of genetic variation in tamoxifen-metabolizing enzymes with overall survival and recurrence of disease in breast cancer patients. Breast Cancer Res Treat 2005;91:249–258.
Wegman P, Vainikka L, Stål O, et al. Genotype of metabolic enzymes and the benefit of tamoxifen in postmenopausal breast cancer patients. Breast Cancer Res 2005;7:R284–R290.
Hayes DF, Stearns V, Rae J, Flockhart D; Consortium on Breast Cancer Pharmacogenomics. A model citizen? Is tamoxifen more effective than aromatase inhibitors if we pick the right patients? J Natl Cancer Inst 2008;100:610–613.
Schroth W, Goetz MP, Hamann U, et al. Association between CYP2D6 polymorphisms and outcomes among women with early stage breast cancer treated with tamoxifen. JAMA 2009;302:1429–1436.
AmpliChip CYP450 Test for in vitro diagnostics use [package insert]. Roche Molecular Systems I: Branchburg, NJ, Catalog Number 04591445001-06. 2006.
Luminex xTAB CYP2D6 kit v.3. http://www.luminexcorp.com/prod/groups/public/documents/lmnxcorp/306-cyp2d6-brochure.pdf. Accessed 21 August 2012.
Autogenomics. INFINITI CYP450 2D6I Assay application notes, 16 April 2009. http://www.autogenomics.com/pharma_2D6.php. Accessed 28 September 2010.
Molecular Pathology Checklist. College of American Pathologists. http://www.cap.org/apps/docs/education/lapaudio/pdf/052009_Presentation.pdf. 2010. Accessed 27 October 2010.
Lubin IM, McGovern MM, Gibson Z, et al. Clinician perspectives about molecular genetic testing for heritable conditions and development of a clinician-friendly laboratory report. J Mol Diagn 2009;11:162–171.
Centers for Disease Control. Genetic Testing Reference Materials Coordination Program (GeT-RM) – Home. http://www.n.cdc.gov/dls/genetics/rmmaterials/default.aspx. Accessed 31 December 2008.
Pratt VM, Zehnbauer B, Wilson JA, et al. Characterization of 107 genomic DNA reference materials for CYP2D6, CYP2C19, CYP2C9, VKORC1, and UGT1A1: a GeT-RM and Association for Molecular Pathology collaborative project. J Mol Diagn 2010;12:835–846.
Decisive Diagnostics. http://www.decisivedx.com/products/. Accessed 29 June 2012.
CAP Reference Resources and Publications, Genetics. http://www.cap.org/apps/cap.portal?_nfpb=true&cntvwrPtlt_actionOverride=%2Fportlets%2FcontentViewer%2Fshow&_windowLabel=cntvwrPtlt&cntvwrPtlt%7BactionForm.contentReference%7D=committees%2Fgenetics%2Fgenetics_index.html&_state=maximized&_pageLabel=cntvwr. Accessed 19 February 2011.
Bradford LD . CYP2D6 allele frequency in European Caucasians, Asians, Africans and their descendants. Pharmacogenomics 2002;3:229–243.
Xie HG, Kim RB, Wood AJ, Stein CM . Molecular basis of ethnic differences in drug disposition and response. Annu Rev Pharmacol Toxicol 2001;41:815–850.
Zhou SF . Polymorphism of human cytochrome P450 2D6 and its clinical significance: Part I. Clin Pharmacokinet 2009;48:689–723.
Sistonen J, Sajantila A, Lao O, Corander J, Barbujani G, Fuselli S . CYP2D6 worldwide genetic variation shows high frequency of altered activity variants and no continental structure. Pharmacogenet Genomics 2007;17:93–101.
Author information
Authors and Affiliations
Consortia
Corresponding author
Supplementary information
Supplementary Table S1.
(DOC 179 kb)
Appendix A.
(DOC 25 kb)
Rights and permissions
About this article
Cite this article
Lyon, E., Gastier Foster, J., Palomaki, G. et al. Laboratory testing of CYP2D6 alleles in relation to tamoxifen therapy. Genet Med 14, 990–1000 (2012). https://doi.org/10.1038/gim.2012.108
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gim.2012.108
Keywords
This article is cited by
-
Relationship between CYP2D6 genotype, activity score and phenotype in a pediatric Thai population treated with risperidone
Scientific Reports (2021)
-
Impact of variable CYP genotypes on breast cancer relapse in patients undergoing adjuvant tamoxifen therapy
Cancer Chemotherapy and Pharmacology (2014)
-
CYP2D6 Genotype and Tamoxifen Activity: Understanding Interstudy Variability in Methodological Quality
Clinical Pharmacology & Therapeutics (2013)