Abstract
Purpose: Alagille syndrome is an autosomal dominant condition characterized by a paucity of interlobular bile ducts and chronic cholestasis, cardiac disease, skeletal abnormalities, ocular abnormalities, and characteristic facies. Most cases harbor a mutation in JAG1. We describe a large consanguineous family with five individuals affected with an Alagille-like syndrome that appears to be autosomal recessive. Our objective was to characterize the disorder clinically and determine whether affected individuals had inherited a mutation in JAG1.
Methods: Clinical data were obtained through questioning and patient chart review. Linkage analysis using microsatellite markers was used to assess the possibility of a JAG1 mutation.
Results: The clinical phenotype of patients was not entirely consistent with classic Alagille syndrome. All affected individuals had neonatal cholestasis with intrahepatic bile duct paucity, with three having pulmonary stenosis, but the presentation was unusually uniform and severe in childhood. There was no evidence of posterior embryotoxon or vertebral anomalies. Cardiac abnormalities were inconsistent between patients. Most significantly, the pedigree suggested an autosomal recessive form of inheritance. Linkage analysis excluded a mutation in JAG1.
Conclusions: We have identified a kindred with an Alagille-like syndrome with an autosomal recessive form of inheritance not caused by a mutation in JAG1.
Similar content being viewed by others
Main
Alagille syndrome, or arteriohepatic dysplasia (AGS) (OMIM #118450), is an autosomal dominant condition first described in 1975.1 It is increasingly recognized as an important and relatively common cause of neonatal jaundice and cholestasis in older children, with an estimated incidence of 1 in 100,000 births.2 The diagnosis of AGS has traditionally been based on the finding of paucity of the interlobular bile ducts associated with at least three of five major features: chronic cholestasis, cardiac disease, skeletal abnormalities (abnormal “butterfly” vertebrae), ocular abnormalities including posterior embryotoxon and retinal pigmentary changes, and a characteristic facial phenotype (broad forehead, deep set eyes, pointed mandible, and bulbous tip of the nose).3
A study conducted by Dhorne-Pollet et al.4 concluded that penetrance for AGS is 94% and that 15% of cases are sporadic. Furthermore, the phenotypic findings associated with AGS are highly variable, ranging from an apparently normal phenotype to severe cases in which liver failure requires transplantation.5 Within a family, one affected person may present with life-threatening congenital heart disease while another shows only subtle abnormalities.6
In 1997, the phenotypic spectrum of clinical AGS was shown to result from defects in a single gene, Jagged1 (JAG1), which has been mapped to chromosome 20p12.7 Li et al.8 demonstrated four different mutations in four affected families. Subsequent reports describe a large number of heterogeneous mutations in patients with a clinical diagnosis of AGS that include whole gene deletions, and frame shift, missense, splice site, and nonsense mutations.7,9,10 The JAG1 locus encodes a ligand for the Notch receptor, which is a key component of an intercellular signaling pathway important in embryonic development.11 Oda et al.12 demonstrated that AGS is caused by mutations that result in haploinsufficency of JAG1.
Recently, heterogeneity for this disorder was confirmed by McDaniell et al.,13 who found NOTCH2 mutations segregating in two of 11 JAG1 mutation-negative probands with AGS. The mechanism by which JAG1 or NOTCH2 defects cause AGS is not currently understood. However, Jagged/Notch interactions are known to be critical for determining cell fates and are essential to normal embryonic development.11
We have identified a large consanguineous Canadian First Nations Miqmaq family with five individuals from two separate sibships who presented with neonatal cholestasis and had features that were initially described as being consistent with AGS. Other nonsyndromic etiologies, such as Zellweger syndrome, were ruled out. However, there were a number of features that were inconsistent with a classic diagnosis of AGS. The presentation was unusually consistent; the clinical course of the liver disease in three of the affected patients was severe; posterior embryotoxon and vertebral anomalies were absent and cardiac abnormalities were inconsistent between patients. Although the affected children have broad foreheads and pointed chins, this was not inconsistent with their ethnic background. Most significantly, the inheritance pattern and the consanguineous nature of the kindred was more in keeping with an autosomal recessive than autosomal dominant condition. These findings are consistent with the genetic heterogeneity of AGS and raise the possibility that the affected members of this kindred have inherited a mutation in a gene other than JAG1 or NOTCH2, which have shown autosomal dominant inheritance in all cases described to date. Indeed, the studies presented here have used linked polymorphic DNA markers to follow the segregation of JAG1 alleles within this family and have shown that the disease does not result from a mutation in JAG1. Other genes in the Jagged/Notch pathway, including NOTCH2, are likely candidates.
MATERIALS AND METHODS
Pedigree and sample acquisition
This study was approved by the Research Ethics Board from both the Capital District Health Authority and IWK health centers, and all patients participated with informed consent.
Through consultation with family members, a seven-generation pedigree was constructed (Fig. 1). Tissue samples were collected with informed parental consent from a total of five affected individuals. These included peripheral blood (10 mL of ethylenediaminetetraacetic acid) from three living children and archived paraffin-embedded tissue from two who were deceased. Collection of peripheral blood (20 mL of ethylenediaminetetraacetic acid) from 23 unaffected family members was facilitated by a community health nurse who coordinated a meeting of the family and research team at the local community center.
Clinical assessment
Family members who consented to give blood were photographed to assess facial features. Unaffected individuals were questioned about relevant medical history including a history of jaundice, other liver disease, cardiac malformations/murmurs, renal anomalies, hypothyroidism, and eye disorders. Blood samples were analyzed for both liver transaminases and bilirubin.
Genotyping
DNA was extracted from peripheral blood lymphocytes according to a standard salting-out protocol. Each sample was genotyped by polymerase chain reaction (PCR) at four polymorphic microsatellite loci that are located closely flanking to the JAG1 gene. These include D20S189 and D20S894, which are located approximately 880 kb and 82 kb proximal to JAG1, as well as D20S160 and D20S162, which are 120 kb and 581 kb distal to the gene, respectively.8
PCR conditions were the same for all reactions, with a 100-ng genomic DNA template, 20 mM Tris HCl (pH 8.4), 50 mM KCl, 1.5 mM MgCl2, 200 μM deoxyribonucleoside triphosphate (each), 1 μM forward and 1μM reverse primer, 1 unit Taq DNA polymerase (Invitrogen, Burlington, Ontario, Canada), and 0.1 μL alpha 32PdCTP (GE Healthcare, Baie d′Urfé, Quebec, Canada) in a total volume of 15 μL. The PCR parameters were 5 minutes denaturation at 96°C, 30 cycles of 96°C for 30 seconds, 55°C for 30 seconds, and 72°C for 30 seconds, and a final extension at 72°C for 10 minutes. The reaction was stopped with 10 μL of “stop solution” (95% formamide, 10 mM NaOtl, 0.25% bromophenol blue, 0.25% xylene cyanol), DNA was denatured for 10 minutes at 65°C, cooled on ice, and then separated electrophoretically on a 6% sequencing gel at 80 W for 3 hours. Gels were dried and exposed autoradiographically at room temperature overnight. PCR primers used to amplify microsatellite alleles are listed in Table 1.
RESULTS
Clinical features
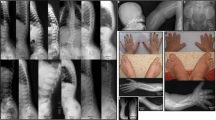
The clinical features of the five affected individuals have been summarized in Table 2. All had documented paucity of the intrahepatic bile ducts, three had cardiac findings, and of the four who were investigated, all had renal tubular acidosis and generalized aminoaciduria. In the sibship with two affected brothers, both had congenital hypothyroidism and elevated sweat chloride values. Neither had any of the 31 common cystic fibrosis mutations found in the Maritime Provinces population, and neither developed a chronic cough or had pancreatic insufficiency. One brother developed juvenile dermatomyositis. Facial features of the brothers are shown in Figure 2. Documented gamma-glutamyl transferase (GGT) levels were not elevated, which was surprising given the history of cholestasis. Thirty-one individuals (mostly first- and second-degree relatives of the affecteds) in the extended family were investigated, including both mothers of the affected children. Unfortunately, neither father of an affected child consented to an assessment, but neither was reported by their spouse to have had any of the medical problems typically seen in AGS. Of the extended family that was investigated, none had any indication of being affected with an Alagille-like syndrome. None had elevations in liver transaminases or bilirubin, and by report, none had hypothyroidism or cardiac disease.
Affected children. Each child is labeled according to the individual's position in the pedigrees depicted in Figure 4.
Pedigree
Figure 1 depicts a seven-generation pedigree of the family described here. Multiple affected individuals were identified in two separate sibships. In both cases, there was no history of the disease. The family is highly consanguineous such that all four parents of the two sibships are interrelated. This pedigree is more in keeping with an autosomal recessive mode of inheritance than with autosomal dominant and therefore raises the possibility that the liver disease in this kindred is not associated with JAG1.
Microsatellite analysis
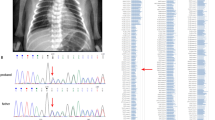
To further assess whether a mutation in JAG1 was responsible for disease in these children, individual JAG1 alleles were defined by microsatellite haplotypes and their segregation through the family was followed to determine if all affected individuals had inherited at least one JAG1 allelle in common that was not inherited by any of the unaffected individuals. Figure 3 shows the position of each marker loci with respect to the JAG1 locus. Figure 4 shows the individual haplotypes deducted from genotype data at four marker loci linked to JAG1, for three affected individuals, their unaffected siblings and their mothers. Genotyping was unsuccessful for two patients (Fig. 4A, II-1 and II-3), where only paraffin-embedded tissue was available. Neither father donated a blood sample, but paternal haplotypes were deduced where possible, based on the haplotypes of the children. Figure 4A shows that the affected child, II-4, inherited the same maternal haplotype as two unaffected siblings, II-5 and II-7. The remaining two unaffected siblings, II-6 and II-9, inherited the alternate maternal haplotype. Similarly, patient II-4 inherited the same paternal haplotype as her unaffected siblings, II-5 and II-7. These data rule out the possibility that a disease-causing JAG1 mutation was inherited from either parent. This conclusion was supported further by haplotype analysis of the second sibship, illustrated in Figure 4B. Although three of the four markers were not highly informative, D20S160 showed four distinct parental alleles. The two affected brothers, II-1 and II-2, inherited different D20S160 alleles from both parents. Because this marker locus is located <100 kb distal to JAG1, they segregate together with a probability of >99.9%. Therefore, a mutation in JAG1 is almost certainly not the cause of disease in these affected brothers.
Haplotype data for affected individuals and their parents. Pedigrees showing sibships with three and two affected children are illustrated in Figure 3A and B, respectively. Haplotyes are presented beneath individual symbols as allele size (number of base pairs) in the order that they occur on the chromosome. To the right of the pedigree is an autoradiogram showing alleles for key individuals at locus D20S160, which is the marker locus located closest to JAG1.
DISCUSSION
We report a family with an Alagille-like syndrome, inherited in an autosomal recessive manner, that is not linked to the JAG1 locus on 20p12. All five affected children studied presented with significant cholestasis in infancy. Liver biopsies revealed a paucity of the bile ducts, consistent with AGS in all children. Two children received a liver transplant, and one died before transplantation could be performed, illustrating the fairly uniform severity of the liver disease in this family. Two of the children had pulmonary valve stenosis, and one had a mild peripheral left pulmonary stenosis. None of the children had ocular findings seen in AGS, and none had documented vertebral anomalies. The faces of the affected individuals were typical for their ethnic background and were not thought to allow for a diagnosis of AGS by two clinical geneticists. Four children were documented to have renal tubular acidosis and generalized aminoaciduria, requiring treatment. The remaining child died before testing was complete. No other renal defects were described; in particular, there were no cysts nor was there renal dysplasia. In addition in the sibship of two brothers, both had congenital hypothyroidism and elevated sweat chloride values. Although all five children had liver involvement, four were documented to have renal tubular acidosis consistent with the diagnosis of AGS, and several had typical cardiac findings of the condition; the lack of ocular, vertebral, and definitive facial findings in any affected child is unusual in this diagnosis.14
Alagille syndrome is an autosomal dominant condition that is typically variable in its presentation. In contrast, in the family presented here, all affected individuals have presented in a homogeneous fashion, all with infantile cholestasis. It would also be unusual for an autosomal dominant condition to express such a high degree of nonpenetrance. Indeed, Kamath et al.14 reported that 31% of affected relatives of AGS probands had liver disease. Based on our review of questionnaires, facial photographs and biochemical analysis of members of the extended family, no other family member has been diagnosed with elevated transaminases, cholestasis, renal anomalies or congenital cardiac anomalies. Although not all relatives underwent a complete physical assessment, it is unlikely that significant issues such as cardiac disease would be under reported given the free access to health care in Canada and the family's relatively close proximity to secondary and tertiary medical care.
The high degree of consanguinity and the consistency of disease presentation in affected children in this kindred are strongly suggestive that it is the result of an autosomal recessive disorder that is likely the result of a founder mutation. Autosomal recessive conditions related to hepatic dysfunction have previously been described in Native American First Nations communities. For example, North American Indian childhood cirrhosis is an autosomal recessive intrahepatic cholestasis syndrome found in Ojibway-Cree from Northwest Quebec.15 It is characterized by transient neonatal jaundice, progressing to biliary cirrhosis with pathological findings of bile duct involvement, with fibrosis around portal bile ducts, and elevated GGT and alkaline phosphatase levels. In this extended kindred, the condition is caused by a founder mutation, R565W, in the cirhin (FLJ14728) gene.16 Our Native American family does not appear to have this condition for several reasons. Although both conditions cause cirrhosis, there are differences in liver pathology: abnormal function of bile ducts with periportal fibrosis in North American Indian childhood cirrhosis is quite different from the bile duct paucity seen in our kindred. The North American Indian childhood cirrhosis kindred has an elevated GGT and no renal involvement, whereas our kindred has normal GGT levels and significant renal involvement. Furthermore, although both groups are of Native American descent (Canadian First Nations), they are different tribes that do not share a common language. Geographically, they are separated by thousands of miles. Interestingly, among Canadian First Nations patients with pyruvate carboxylase deficiency, 11 of 13 affected children were of Ojibwa or Cree descent and had a homozygous founder mutation, c.1828G-A in the pyruvate carboxylase gene.17 However, the two affected members of a Miqmaq kindred in Nova Scotia, had a homozygous mutation, c.2229G-T, not seen in the other tribes, illustrating heterogeneity between the Maritime Miqmaq population and other tribes in Canada. It is therefore not surprising that this Alagille-like condition has not been identified in other Canadian First Nations Groups.
Several other autosomal recessive conditions with cholestasis and renal tubular acidosis have been described. Nephronophthisis type 2 is an autosomal recessive condition characterized by severe renal failure and acidosis, hypertension, and polyuria, in which most children have developed renal failure by 2 years of age.18 Uncommonly, patients with this condition have developed cirrhosis. Given that the renal involvement in our kindred did not evolve into renal failure in any individual and that liver dysfunction was present in all children affected with this Alagille-like condition, this diagnosis is unlikely in our kindred. Individuals with the autosomal recessive disorder glutaric aciduria type II, have also been described as having renal tubular acidosis and/or cholestasis,19 but this condition is associated with other features, such as a typical organic acid profile and clinical features that our patients did not exhibit, making this diagnosis also unlikely.
Approximately 94% of individuals with a clinical diagnosis of AGS have been shown to harbor a mutation or deletion in JAG1.7–10,12,20 Li et al.7,20 described a large interstitial deletion that included JAG1 and four distinct frame shift mutations within conserved regions of the gene. Similarly, Oda et al.12 and/or Yuan et al.7 identified frame shift, splice site, and nonsense mutations. Similar heterogeneity was reported in a large study of 233 Alagille syndrome patients reported by Spinner et al.9 In all cases, the mutations caused gross alterations to the protein and were present in a heterozygous form. Accordingly, Oda et al.12 concluded that AGS results from haploinsufficiency of JAG1, which is consistent with its autosomal dominant form of inheritance. Heterozygosity and autosomal dominant inheritance were also found with the two recently reported AGS families with NOTCH2 mutations.13 The work described here negates JAG1 as the genetic defect in this family. The affected brothers did not share a single common haplotype at this region. However, despite the apparent autosomal recessive mode of inheritance, NOTCH2 mutations have not been ruled out. It is conceivable that a minor alteration producing a partially functional protein, which is expected to be homozygous in this highly consanguineous pedigree, could result in autosomal recessive inheritance. Indeed, preliminary linkage analysis does not rule out NOTCH2 as a candidate gene. Currently, work is under way to assess this and other genes in the Jagged/Notch pathway as candidate genes.
In our search for the genetic defect, it is important to consider the possibility that the two families within this kindred had separate medical conditions. However, given that all affected individuals had interlobular bile duct paucity, which is rare, and there was sibling recurrence in both branches of this family, it is likely that the disease in each kindred is the same genetic entity. The fact that all affected children are from the same small geographic and ethnic community gives further suggestion that they have the same genetic condition. The affected brothers have additional features, including congenital hypothyroidism and, in one case, juvenile dermatomyositis, that may or may not be related to their Alagille-like condition. It is possible that this is an expression of variability in the genetic condition affecting these children. Alternatively, given the high degree of consanguinity, these boys may have inherited a separate autosomal recessive form of hypothyroidism and/or an autosomal recessive or multifactorial autoimmune disorder that was not inherited by the other three affected children. Also, elevated sweat chloride values have been reported in patients with hypothyroidism and may not be a feature of this Alagille-like condition.21 Nevertheless, a mutation in JAG1 as the cause of disease has been ruled out independently for each sibship, and the presentation of affected children with neonatal cholestasis and paucity of the intrahepatic bile ducts on the initial liver biopsy strongly support an autosomal recessive disorder rather than a highly variable autosomal dominant disorder, as is characteristic of classic AGS.
In summary, we have identified a family with an AGS-like condition with an autosomal recessive mode of inheritance that is not due to a mutation in JAG1. This information has significant implications for genetic counseling for this and other AGS families. Given the nature of the population, we expect that there are many other individuals at risk of having affected children, and we are currently unable to offer carrier testing or prenatal diagnosis. Determining the specific gene involved will help us clinically and will lead to a better understanding of the pathophysiology of this condition and perhaps lead to better therapies in the future. NOTCH2 and other genes involved in this developmental pathway are obvious candidate genes responsible for disease in this kindred.
References
Alagille D, Odievre M, Gautier M, Dommergues JP . Hepatic ductular hypoplasia associated with characteristic facies, vertebral malformations, retarded physical, mental and sexual development and cardiac murmur. J Pediatr 1975; 86: 63–71.
Piccoli DA, Witzleben CL . Disorders of the intrahepatic bile ducts. In: Walker WA, Durie PR, Hamilton JR, Walker-Smith JA, et al, editors. Pediatric gastrointestinal disease: pathophysiology, diagnosis, management, volume two. St. Louis: Mosby, 1996; 1362–1384.
Emerick KM, Rand EB, Goldmuntz E, Krantz ID, et al. Features of Alagille syndrome in 92 patients: frequency and relation to prognosis. Hepatology 1999; 29: 822–829.
Dhorne-Pollet S, Deleuze JF, Hadchouel M, Bonaiti-Pellie C . Segregation analysis of Alagille syndrome. J Med Genet 1994; 31: 453–457.
Crosnier C, Attie-Bitach T, Encha-Razavi F, Audollent S, et al. JAGGED1 gene expression during human embryogenesis elucidates the wide phenotypic spectrum of Alagille syndrome. Hepatology 2000; 32: 574–581.
LaBrecque DR, Mitros FA, Nathan RJ, Romanchuk KG, et al. Four generations of arteriohepatic dysplasia. Hepatology 1982; 2: 467–474.
Yuan ZR, Kohsaka T, Ikegaya T, Suzuki T, et al. Mutational analysis of the Jagged 1 gene in Alagille syndrome families. Hum Mol Genet 1998; 7: 1363–1369.
Li L, Krantz ID, Deng Y, Genin A, et al. Alagille syndrome is caused by mutations in human Jagged 1, which encodes a ligand for Notch 1. Nat Genet 1997; 16: 243–151.
Spinner NB, Colliton RP, Crosnier C, Krantz ID, et al. Jagged 1 mutations in Alagille syndrome. Hum Mutat 2001; 17: 18–33.
Warthen DM, Moore EC, Kamath BM, Morrissette JJ, et al. Jagged 1 (JAG1) mutations in Alagille syndrome: increasing the mutation detection rate. Hum Mutat 2006; 27: 436–443.
Artavanis-Tsakonas S . Alagille syndrome—a notch up for the Notch receptor. Nat Genet 1997; 16: 212–213.
Oda T, Elkahloun AG, Meltzer PS, Chandrasekharappa SC . Identification and cloning of the human homolog (JAG1) of the rat Jagged1 gene from the Alagille syndrome critical region at 20p12. Genomics 1997; 43: 376–379.
McDaniell R, Warthen DM, Sanchez-Lara PA, Pai A, et al. NOTCH2 mutations cause Alagille syndrome, a heterogeneous disorder of the notch signaling pathway. Am J Hum Genet 2006; 79: 169–173.
Kamath BM, Bason L, Piccoli DA, Krantz ID, et al. Consequences of JAG1 mutations. J Med Genet 2003; 40: 891–895.
Drouin E, Russo P, Tuchweber B, Mitchel G, et al. North American Indian cirrhosis in children: a review of 30 cases. J Pediatr Gastroenterol Nutr 2000; 31: 395–404.
Chagnon P, Michaud J, Mitchell G, Mercier J, et al. A missense and mutation (R565W) in Cirhin (FLJ14728) in North American Indian childhood cirrhosis. Am J Hum Genet 2002; 71: 1443–1449.
Robinson BH, Oei J, Sherwood WG, Applegarth D, et al. The molecular basis for the two different clinical presentations of classical pyruvate carboxylase deficiency. Am J Hum Genet 1984; 36: 283–294.
Gagnadoux MF, Bacri JL, Broyer M, Habib R . Infantile chronic tubulo-interstitial nephritis with cortical microcysts: variant of nephronophthisis or new disease entity?. Pediatr Nephrol 1989; 3: 50–55.
Scriver CR, Beaudet AL, Sly WS, Valle D, editors. The metabolic and molecular basis of inherited disease, volume II, 8th ed. New York: McGraw-Hill, 2001: 2359–2363.
Li PH, Shu SG, Yang CH, Lo FC, et al. Alagille syndrome with interstitial 20P deletion derived from maternal ins(7;20). Am J Med Genet 1996; 63: 537–541.
Madoff L . Elevated sweat chlorides and hypothyroidism. J Pediatr 73: 244–246, 1968; Aug.
Acknowledgements
This work was supported by grants from the Capital District Health Authority and the IWK Health Centre in Halifax.
Author information
Authors and Affiliations
Corresponding author
Additional information
Disclosure: The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Dyack, S., Cameron, M., Otley, A. et al. An autosomal recessive form of Alagille-like syndrome that is not linked to JAG1. Genet Med 9, 544–550 (2007). https://doi.org/10.1097/GIM.0b013e318133a802
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1097/GIM.0b013e318133a802
Keywords
This article is cited by
-
Alagille syndrome: pathogenesis, diagnosis and management
European Journal of Human Genetics (2012)