Abstract
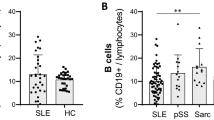
Systemic lupus erythematosus (SLE) is a systemic autoimmune disease that is known to be associated with polyclonal B-cell hyper-reactivity. B-cell receptor (BCR) has a central role in B-cell development, activation, survival and apoptosis, and thus is a critical component of the regulation of both protective and autoreactive B cells. In this study, we applied multiplex PCR and Illumina high-throughput sequencing to study the composition and variation of the BCRs in peripheral blood mononuclear cells from SLE patients and healthy donors (NC). We found that SLE group displayed significantly shorter CDR3 average length (14.86±0.76aa vs 15.70±0.43aa), more arginine percentage of CDR3 amino acids (7.57±0.20% vs 7.32±0.19%) and poorer immunological diversity than the healthy ones. CDR3 sequence YGMDV present in all SLE samples may provide more information in generating more effective B-cell targeted diagnosis/therapies strategies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 digital issues and online access to articles
$119.00 per year
only $19.83 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Taylor EB, Ryan MJ . Understanding mechanisms of hypertension in systemic lupus erythematosus. Ther Adv Cardiovasc Dis 2016, e-pub ahead of print 15 March 2016 doi:10.1177/1753944716637807.
Shlomchik MJ, Craft JE, Mamula MJ . From T to B and back again: positive feedback in systemic autoimmune disease. Nat Rev Immunol 2001; 1: 147–153.
Meffre E, Wardemann H . B-cell tolerance checkpoints in health and autoimmunity. Curr Opin Immunol 2008; 20: 632–638.
Paran D, Naparstek Y . Is B cell-targeted therapy effective in systemic lupus erythematosus? Isr Med Assoc J 2015; 17: 98–103.
Sanz I, Lee FE . B cells as therapeutic targets in SLE. Nat Rev Rheumatol 2010; 6: 326–337.
Bakshi J, Ismajli M, Rahman A . New therapeutic avenues in SLE. Best Pract Res Clin Rheumatol 2015; 29: 794–809.
Jung D, Giallourakis C, Mostoslavsky R, Alt FW . Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Annu Rev Immunol 2006; 24: 541–570.
Calis JJ, Rosenberg BR . Characterizing immune repertoires by high throughput sequencing: strategies and applications. Trends Immunol 2014; 35: 581–590.
Mori A, Deola S, Xumerle L, Mijatovic V, Malerba G, Monsurrò V . Next generation sequencing: new tools in immunology and hematology. Blood Res 2013; 48: 242–249.
Weinstein JA, Jiang N, White RA 3rd, Fisher DS, Quake SR . High-throughput sequencing of the zebrafish antibody repertoire. Science 2009; 324: 807–810.
Robins HS, Campregher PV, Srivastava SK, Wacher A, Turtle CJ, Kahsai O et al. Comprehensive assessment of T-cell receptor beta-chain diversity in alphabeta T cells. Blood 2009; 114: 4099–4107.
Freeman JD, Warren RL, Webb JR, Nelson BH, Holt RA . Profiling the T-cell receptor beta-chain repertoire by massively parallel sequencing. Genome Res 2009; 19: 1817–1824.
Warren RL, Freeman JD, Zeng T, Choe G, Munro S, Moore R et al. Exhaustive T-cell repertoire sequencing of human peripheral blood samples reveals signatures of antigen selection and a directly measured repertoire size of at least 1 million clonotypes. Genome Res 2011; 21: 790–797.
Wu D, Sherwood A, Fromm JR, Winter SS, Dunsmore KP, Loh ML et al. High-throughput sequencing detects minimal residual disease in acute T lymphoblastic leukemia. Sci Transl Med 2012; 4: 134ra63.
Wu D, Emerson RO, Sherwood A, Loh ML, Angiolillo A, Howie B et al. Detection of minimal residual disease in B lymphoblastic leukemia by high-throughput sequencing of IGH. Clin Cancer Res 2014; 20: 4540–4548.
Robins H . Detecting and monitoring lymphoma with high-throughput sequencing. Oncotarget 2011; 2: 287–288.
Sui W, Hou X, Zou G, Che W, Yang M, Zheng C et al. Composition and variation analysis of the TCR beta-chain CDR3 repertoire in systemic lupus erythematosus using high-throughput sequencing. Mol Immunol 2015; 67: 455–464.
Meyer EH, Hsu AR, Liliental J, Löhr A, Florek M, Zehnder JL et al. A distinct evolution of the T-cell repertoire categorizes treatment refractory gastrointestinal acute graft-versus-host disease. Blood 2013; 121: 4955–4962.
Grimaldi CM, Hicks R, Diamond B . B cell selection and susceptibility to autoimmunity. J Immunol 2005; 174: 1775–1781.
Sfikakis PP, Karali V, Lilakos K, Georgiou G, Panayiotidis P . Clonal expansion of B-cells in human systemic lupus erythematosus: evidence from studies before and after therapeutic B-cell depletion. Clin Immunol 2009; 132: 19–31.
Jacobi AM, Diamond B . Balancing diversity and tolerance: lessons from patients with systemic lupus erythematosus. J Exp Med 2005; 202: 341–344.
Mietzner B, Tsuiji M, Scheid J, Velinzon K, Tiller T, Abraham K et al. Autoreactive IgG memory antibodies in patients with systemic lupus erythematosus arise from nonreactive and polyreactive precursors. Proc Natl Acad Sci USA 2008; 105: 9727–9732.
Shi B, Yu J, Ma L, Ma Q, Liu C, Sun S et al. Short-term assessment of BCR repertoires of SLE patients after high dose glucocorticoid therapy with high-throughput sequencing. Springerplus 2016; 5: 75.
Yurasov S, Wardemann H, Hammersen J, Tsuiji M, Meffre E, Pascual V et al. Defective B cell tolerance checkpoints in systemic lupus erythematosus. J Exp Med 2005; 201: 703–711.
Wardemann H, Yurasov S, Schaefer A, Young JW, Meffre E, Nussenzweig MC . Predominant autoantibody production by early human B cell precursors. Science 2003; 301: 1374–1377.
Aguilera I, Melero J, Nuñez-Roldan A, Sanchez B . Molecular structure of eight human autoreactive monoclonal antibodies. Immunology 2001; 102: 273–280.
Meffre E, Milili M, Blanco-Betancourt C, Antunes H, Nussenzweig MC, Schiff C . Immunoglobulin heavy chain expression shapes the B cell receptor repertoire in human B cell development. J Clin Invest 2001; 108: 879–886.
Meffre E, Schaefer A, Wardemann H, Wilson P, Davis E, Nussenzweig MC . Surrogate light chain expressing human peripheral B cells produce self-reactive antibodies. J Exp Med 2004; 199: 145–150.
Elhanati Y, Sethna Z, Marcou Q, Callan CG Jr, Mora T, Walczak AM . Inferring processes underlying B-cell repertoire diversity. Philos Trans R Soc Lond B Biol Sci 2015; 370: 1676.
Hoehn KB, Fowler A, Lunter G, Pybus OG . The diversity and molecular evolution of B-cell receptors during infection. Mol Biol Evol 2016; 33: 1147–1157.
Breden F, Lepik C, Longo NS, Montero M, Lipsky PE, Scott JK . Comparison of antibody repertoires produced by HIV-1 infection, other chronic and acute infections, and systemic autoimmune disease. PLoS One 2011; 6: e16857.
Magurran A E . Biological diversity. Curr Biol 2005; 15: 116–118.
Yin L, Hou W, Liu L, Cai Y, Wallet MA, Gardner BP et al. IgM repertoire biodiversity is reduced in HIV-1 infection and systemic lupus erythematosus. Front Immunol 2013; 4: 373.
Dörner T, Giesecke C, Lipsky PE . Mechanisms of B cell autoimmunity in SLE. Arthritis Res Ther 2011; 13: 243.
Tipton CM, Fucile CF, Darce J, Chida A, Ichikawa T, Gregoretti I et al. Diversity, cellular origin and autoreactivity of antibody-secreting cell population expansions in acute systemic lupus erythematosus. Nat Immunol 2015; 16: 755–765.
Liu X, Zhang W, Zeng X, Zhang R, Du Y, Hong X et al. Systematic comparative evaluation of methods for investigating the TCRβ repertoire. PLoS One 2016; 11: e0152464.
Han Y, Li H, Guan Y, Huang J . Immune repertoire: A potential biomarker and therapeutic for hepatocellular carcinoma. Cancer Lett 2016; 379: 206–212.
Han Y, Liu X, Wang Y, Wu X, Guan Y, Li H et al. Identification of characteristic TRB V usage in HBV-associated HCC by using differential expression profiling analysis. Oncoimmunology 2015; 4: e1021537.
Zhang W, Du Y, Su Z, Wang C, Zeng X, Zhang R . IMonitor: A robust pipeline for TCR and BCR repertoire analysis. Genetics 2015; 201: 459–472.
Acknowledgements
This work was supported by funds received from Science and Technology Plan of Shenzhen, Guangdong (No. JCYJ20160422150329190), the fund received from National Natural Science Foundation of China (No. 81671596).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on Genes and Immunity website
Rights and permissions
About this article
Cite this article
Liu, S., Hou, X., Sui, W. et al. Direct measurement of B-cell receptor repertoire’s composition and variation in systemic lupus erythematosus. Genes Immun 18, 22–27 (2017). https://doi.org/10.1038/gene.2016.45
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gene.2016.45
This article is cited by
-
B-Cell Receptor Repertoire: Recent Advances in Autoimmune Diseases
Clinical Reviews in Allergy & Immunology (2024)
-
TRUST4: immune repertoire reconstruction from bulk and single-cell RNA-seq data
Nature Methods (2021)
-
Involvement of transcribed lncRNA uc.291 and SWI/SNF complex in cutaneous squamous cell carcinoma
Discover Oncology (2021)
-
Landscape of B cell immunity and related immune evasion in human cancers
Nature Genetics (2019)