Abstract
G-quadruplexes (G4) are secondary nucleic acid structures that have been associated with genomic instability and cancer progression. When present in the promoter of some oncogenes, G4 structures can affect gene regulation and, hence, represent a possible therapeutic target. In this study, RNA-Seq was used to explore the effect of a G4-binding anthraquinone derivative, named AQ1, on the whole-transcriptome profiles of two common cell models for the study of KIT pathways; the human mast cell leukemia (HMC1.2) and the canine mast cell tumor (C2). The highest non-cytotoxic dose of AQ1 (2 µM) resulted in 5441 and 1201 differentially expressed genes in the HMC1.2 and C2 cells, respectively. In both cell lines, major pathways such as cell cycle progression, KIT- and MYC-related pathways were negatively enriched in the AQ1-treated group, while other pathways such as p53, apoptosis and hypoxia-related were positively enriched. These findings suggest that AQ1 treatment induces a similar functional response in the human and canine cell models, and provide news insights into using dogs as a reliable translational model for studying G4-binding compounds.
Similar content being viewed by others
Introduction
Targeted therapy is a novel strategy for cancer treatment that aims to destroy cancer cells with more precision and, potentially, fewer side effects1. For the discovery of clinical agents targeting oncogenic signaling pathways, it is important to define the compound’s specificity towards the target molecular pathway. In this respect, the field of cancer genetics and genomics is rapidly evolving, particularly focusing on targeted pathways that are critically linked to a malignant behavior2. Currently, multiple compounds are being used to treat cancer and many more are being tested in clinical trials3,4,5.
Canine/human comparative oncology is recently playing an increasingly important role in advancing cancer research from basic science, preclinical and clinical perspectives. The acknowledgment of spontaneous cancer development in companion animals and the potential of using these animals in drug development studies and preclinical trials is harnessed by the morphologic, histologic and biological similarities between canine and human spontaneously-occurring malignancies. Dogs’ physical size, accessibility to serial biological sample collections, comparable tumor biology, intact immunity and short survival compared to humans led to their inclusion as a complementary animal model2. Inter-species cancer studies have contributed to gaining new insights into the biology of breast cancer, osteosarcoma, lung and bladder cancer6,7,8,9,10. Further, recent studies on canine B-cell lymphoma demonstrated that recurrent aberrations were correlated with cancer subtype, and the corresponding cytogenetic lesions were also noticed in human patients11,12. Meantime, another study showed that canine melanoma possesses rare mutations in the B-Raf (BRAF) and N-ras (NRAS) proto-oncogenes; however, they present similar gene expression changes to human melanoma within the same downstream activation pathways13. Thus, although the oncogenic driving somatic mutations between the two species may differ, similar upregulation and signaling pathways underscore the translational value of studying tumor biology from a comparative point of view. This concept might support the beginning of the so-called ‘oncological basket’ trials, wherein the response of tumors with shared credentialed biology to a specific targeted therapy are assessed beyond histologic diagnosis2.
In the past decade, several studies have been focusing on guanine-rich stretches found in important genomic regions such as telomeres, centromeres, immunoglobulin switch regions, mutational hot spots, and promoter elements14. These guanine-rich sequences may have the potential to form four-stranded secondary structures called G-quadruplexes (G4) that are stabilized by non-Watson-Crick interactions (Hoogsteen bond15). Several studies clarified that G4 structures are often linked to important DNA regions involved in the regulation of gene transcription and translation16,17. Moreover, studies have shown that several proto-oncogenes like v-myc avian myelocytomatosis viral oncogene homolog (MYC), v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog (KIT), B-cell lymphoma 2 (BCL2), vascular endothelial growth factor (VEGF), platelet-derived growth factor (PDGF) and hypoxia-inducible factor (HIF) bear G4 motifs in their promoters16,18,19,20,21,22,23,24. Specific proteins, which selectively bind G4 structures, have been discovered impinging on the biological roles of G425. These motifs could be efficiently targeted by G4-selective ligands, thus altering their molecular recognition and, consequently, affect their downstream expression25.
More than 55% of the genes in the human genome bear at least one potential G4 motif 1 kb up- or downstream of the transcription starting site (TSS); hence, it is important to carefully assess whether the treatment with G4 binding compounds could affect the transcriptional activity of these genes26. Using the G4-stabilizing molecule TMPyP4, previous studies showed genome-wide effects on transcription in human cells; moreover, relatively high doses of TMPyP4 promoted cell migration and apoptosis in human cell lines27,28,29. Likewise, Halder and colleagues demonstrated that bisquilinium compounds were able to induce quadruplex-specific transcriptomic changes in HeLa S3 cells26. Marchetti et al., in 2018, used the RNA-Seq technology to study the effects of a new G4 binding compound in pancreatic ductal adenocarcinoma30.
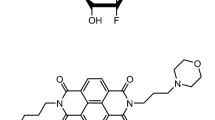
Recently, we demonstrated that an anthraquinone derivative (named AQ1, Supplementary Fig. S1) significantly downregulated KIT mRNA and protein levels in several human cancer cell lines, thereby preventing their proliferation31. Meantime, two KIT G4-forming sequences (d_kit1 and d_kit2) were discovered in the canine genome (Supplementary Fig. S1)32. They showed a high degree of homology with the corresponding human sequences, and preliminary studies were conducted to assess their interaction with AQ133. These studies on G4 structures were executed using a targeted-gene approach; nevertheless, it is of interest to extend the study of AQ1 transcriptional effects to a wider scenario, like the whole-transcriptome. Such an approach is expected to: (1) clarify the AQ1 influence on KIT-related pathways and (2) unveil specific off-targets of AQ1 in species-specific cells. Therefore, in the present study, we investigated - using RNA-Seq- the AQ1-induced effects on the global gene expression in two species-specific KIT-dependent cancer cell lines, in order to identify which cellular pathways are likely to be affected by treatment. For this purpose, we used the HMC1.2 human mast cell leukemia and the C2 canine mast cell tumor cell line, which are two common models for the study of tyrosine kinase inhibitors and, in particular, the proto-oncogene KIT.
Results
Effects of AQ1 on cell cytotoxicity and KIT and BCL2 mRNA levels
We previously demonstrated that AQ1 significantly downregulated KIT and BCL2 oncogenes in human mast cell leukemia HMC1.2, where the major effect of the treatment was observed 12 hours (T12) post-exposure31. Thus, we selected this incubation time to follow the activity of AQ1 on the two cell lines used in this study. Proliferation curves (Supplementary Fig. S2) demonstrated that, after 12 hours of incubation, AQ1 was moderately cytotoxic, with 70–90% cell survival in both the two cell lines at the tested concentration range (0–10 μM). Additionally, quantitative real-time PCR (qPCR) showed a significant dose-dependent reduction of KIT and BCL2 mRNA levels in both HMC1.2 (p < 0.01) and C2 (p < 0.001) cells (Supplementary Fig. S3).
Whole-transcriptome profiling
To account for any possible vehicle- (dimethyl sulfoxide, DMSO) related effects on cell gene expression, we also profiled the transcriptome of untreated cells (treatment- and vehicle-free medium). No differentially expressed genes (DEGs) were detected in the DMSO-treated cells compared with the untreated ones (results not shown); consequently, the DMSO-treated cells dataset was used as control.
The RNA-Seq analysis of the AQ1-treated HMC1.2 cells, compared with DMSO-treated cells, evidenced 3644 and 5441 DEGs (false discovery rate, FDR <0.05 and fold-change, FC >2) at 1 µM and 2 µM AQ1 concentrations, respectively. For both concentrations, more than 60% of the total DEGs were downregulated. In the AQ1-treated C2 cells, the whole-transcriptome analysis identified 397 and 1201 DEGs at 1 µM and 2 µM AQ1 concentration, respectively. Likewise to HMC1.2, more than 70% of these DEGs were downregulated. The whole list of the DEGs for both cell lines is reported in Supplementary Dataset S1 and a list of the common downregulated genes between HMC1.2 and C2 is reported in Supplementary Dataset S2.
Both experimental groups clustered uniquely on a hierarchical clustering plot (Fig. 1a,b), and a Principal Component Analysis (PCA) plot, using the top 100 DEGs, resulted into two distinct clusters of samples with the principal component 1 (PC1) accounting for the major variation between groups (PC1 >98%; Fig. 1c,d). This variation between AQ1-treated and control groups is further visualized using two heatmaps that are shown in Supplementary Fig. S4.
Analysis of intra-population consistency: hierarchical clustering and PCA. (a,b) Hierarchical clustering analyses performed using DESeq2 rlog-normalized RNA-Seq data. (a) HMC1.2 cells, AQ1 (2 µM) vs DMSO. (b) C2 cells, AQ1 (2 µM) vs DMSO. Color code (from white to dark blue) refers to the distance metric used for clustering (dark blue corresponds to the maximum of correlation values). (c,d) Principal Component Analysis (PCA) performed on top 100 differentially expressed genes using ClustVis (Metsalu & Vilo, 2015) tool. Loadings PC1 and PC2 are reported in graph (on x and y-axes). (c) HMC1.2 cells, AQ1 (2 µM) vs DMSO. (d) C2 cells, AQ1 (2 µM) vs DMSO.
Gene set enrichment analysis (GSEA) and protein-protein interactions
Canine C2 and human HMC1.2 cell lines showed similar functional response to AQ1 treatment. Figures 2 and 3 show the most positively- or negatively-enriched cancer hallmarks in the AQ1-treated (2 µM) HMC1.2 (Fig. 2) and C2 cells (Fig. 3). In HMC1.2 cells (Fig. 2), the most negatively-enriched hallmarks were the mTORC1 signaling pathway (FDR = 0.017), the targets of MYC (FDR = 0.026), the G2M cell cycle checkpoint (FDR = 0), and the mitotic spindle (FDR = 0). As shown in Fig. 3, AQ1-treated canine C2 cells show a similar functional response to the human HMC1.2 cells. Further, KEGG pathways analysis (Supplementary Tables S1–S4) identified many pathways that were negatively-enriched in the AQ1-treated samples, in both cell lines34,35,36. For instance, cell-cell adherent junctions (HMC1.2, FDR = 0.009; C2, FDR = 0.043), mismatch repair (HMC1.2, FDR = 0.015; C2, FDR = 0.041), the regulation of actin cytoskeleton (HMC1.2, FDR = 0.012; C2, FDR = 0.036), the DNA replication machinery (FDR = 0.008), the tRNA biosynthesis (FDR = 0.012) and the cell cycle pathway (FDR = 0.014). Figure 4 (panels a,b) shows the downregulation of phosphatidylinositol signaling system (p ≤ 0.001) KEGG pathway in HMC1.2 and C2, respectively. Moreover, the complement and coagulation cascade were the most important KEGG pathway positively enriched in both cells (Fig. 4c,d). Supplementary File 1 report, more in details, the most important pathways identified by KEGG GSEA analysis in HMC1.2 and C2, respectively. This functional similarity was also confirmed by the The Database for Annotation, Visualization and Integrated Discovery (DAVID) tool (data not shown). Also, several important cancer hallmarks, such as P53- and apoptosis-related pathways as well as hypoxia and tumor necrosis factor α (TNFα) pathways related to NF-κβ, were positively enriched (FDR < 0.05) in both cell lines (Figs 2 and 3).
GSEA Enrichment plot (KEGG pathways) negatively and positively enriched in HMC1.2 and C2 cell lines exposed to AQ1 2 μM. The phosphatidylinositol signaling system was negatively enriched by AQ1 in HMC1.2 (panel a) and C2 (panel b). The complement and coagulation cascades pathways was positively enriched by AQ1 in HMC1.2 (panel c) and C2 (panel d). The green curve corresponds to the ES (enrichment score) curve, which is the running sum of the weighted enrichment score obtained from GSEA software, while the normalized enrichment score (NES) and the corresponding FDR are reported within each graph. A permission for the use of the images was obtained by KEGG (Kanehisa Laboratories rif. 180271)34,35,36.
With regard to KIT regulation, the RNA-Seq data evidenced a significant inhibition of multiple downstream processes activated by this proto-oncogene. Figure 5 shows the expression profile (mRNA) of some genes that are part of several KIT downstream pathways such as the Ras/Erk signal transduction, the PI3K signal transduction, the PLC-γ signaling transduction, the Src kinase signal transduction and the JAK/STAT signaling pathways37. In both cell lines, the whole set of target genes were negatively enriched following the AQ1 treatment, except for the canine JAK-STAT pathway.
RNA-Seq fold change (FC) of genes involved in KIT downstream pathways after treatment with AQ1 2 µM. GRB2: Growth Factor Receptor Bound Protein 2. MAPK: Mitogen-Activated Protein Kinase 1. AKT3: AKT Serine/Threonine Kinase 3. mTOR: Mechanistic Target Of Rapamycin Kinase. FYN: FYN Proto-Oncogene, Src Family Tyrosine Kinase. PLCγ2: Phospholipase C Gamma 2. SOS1: SOS Ras/Rac Guanine Nucleotide Exchange Factor 1. BRAF: B-Raf Proto-Oncogene, Serine/Threonine Kinase. LYN: LYN Proto-Oncogene, Src Family Tyrosine Kinase. JAK2: Janus Kinase 2. STAT: Signal Transducer And Activator Of Transcription 1.
To uncover some of the biological consequences the DEGs might have, the top downregulated protein-coding genes were collapsed to their corresponding protein symbols and their interactions were visualized by a protein-protein interaction (PPI) network, using the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING, https://string-db.org/) database. Figure 6 shows the PPI network (p < 0.001) observed in HMC 1.2 cells exposed to 2 µM AQ1. The most negatively-enriched pathways were the transmembrane receptor protein tyrosine kinase signaling pathway (GO.0007169; FDR <0.001), the vascular endothelial growth factor receptor signaling pathway (GO.0048010; FDR <0.001), and the cell surface receptor signaling pathway (GO.0007166; FDR <0.001). Because the STRING database relies largely on curated experimental data, a meaningful PPI for the DEGs of C2 cells was not possibly – likely because little information is available on the function of proteins in dogs.
STRING protein network map of thirty proteins commonly downregulated by AQ1 2 µM in HMC1.2. Colored nodes indicate the individual protein identified. Lines between nodes represent direct and indirect association of proteins. Line colors indicate the type of interaction evidence (light blue: known interaction from curated database; purple: known interaction experimentally determined; green: predicted interaction gene neighborhood; red: predicted interaction gene fusion; blue: predicted interaction gene co-occurrence; yellow: text mining; black: co-expression; lilac: protein homology.
G4 prediction
The Quadbase2/EuQuad tool38,39 was used to mine G4 from RNA-Seq results using the corresponding gene’s Ensembl ID. Figure 7 reports the results obtained searching for the G3 L1–7 motif in ±2 kb TSS of each record. In both species, EuQuad predicted the presence of at least one G4 domain near the TSS for the majority of DEGs (overall >75%). The percentage of DEGs with a predicted G4 domain did not differ between the down- and up-regulated genes (71% vs 85.8% in canine DEGs and 80% vs 76% in human DEGs, respectively).
qPCR confirmation
To cross-validate the RNA-Seq data, twelve genes were tested for their relative gene expression by qPCR. The selected genes belonged to different pathways: KIT activation pathways (RPTOR), epigenetic regulation (HDAC4), apoptosis (BMF), cell cycle (CDKN1A, CDK6), metabolism (CYP1A1, FAXDC2), cell signaling (KRS2, TNFRSF11β), transcription regulation (EGR2), cell morphology (MID1), and tumor oncogenes (EGFR). The qPCR results (Supplementary Figures S5–S7) corroborated the RNA-Seq data analysis. In specific, the downregulation of RPTOR and HDAC4 by AQ1, as well as the upregulation of the pro-apoptotic factor BMF and cell cycle controller CDKN1A were significantly correlated between the qPCR and RNA-Seq data, in both cell lines.
The EuQuad output for each gene selected for qPCR confirmation is shown in Supplementary Table S5. For every gene, the software analyzed around 2 kb before and after the TSS to search for G3 L1-7 motif. HDAC4 was the gene with the highest number of putative G4-forming sequences near the TSS in both species, followed by CDK6 in human and RPTOR in dog. On the contrary, for other genes like canine EGFR, human CDKN1a and human EGR2 the tool did not find G3 L1-7 motifs.
Discussion
DNA is one of the main targets of most anticancer drugs. However, it is rather difficult to achieve a selective effect on particular DNA regions because of the high homogeneity of cellular DNA, mostly occurring in the B form. Interestingly, the possible interactions with specific non-canonical DNA structures may improve the selectivity of low molecular weight compounds towards certain cell processes or signaling pathways. In this scenario, an increasing interest has recently gained targeting the G4 secondary structure of nucleic acids that might affect pathways that are critical for tumor cells growth and proliferation40.
We previously used a targeted approach to assess the effect of AQ1 both in human and canine KIT-dependent cell lines (from two to almost forty-fold inhibition)31,32,33. Results demonstrated a significant inhibition of cell proliferation and downregulation of KIT and BCL2 induced by AQ1 whereas other prototypical oncogenes as hTERT, MYC, and PDGF were not highly modulated by this compound. Moreover, we confirmed the presence of two conserved G4 forming domain in canine cKIT promoter32.
Taking into consideration these evidence, in the present study, we profiled, using RNA-Seq, the effects of AQ1 treatment on the transcriptome of two KIT-dependent cell lines of human and canine origin (HMC1.2 and C2, respectively). The choice of time and doses derived from a preliminary validation step; in particular, the two doses we used were poorly cytotoxic for both cell lines, while the time of exposure (T12) was sufficiently short to attribute the major transcriptional modulation to DNA binding by AQ1 and not to secondary effects that could occur over a longer treatment time. Indeed, the occurrence of such a drawback emerged in previous transcriptional works. For instance, Halder and colleagues (2012) performed a transcriptional profiling with microarray after 48 h of treatment; because of the long-time of treatment, it was not possible to rule out whether or not some or many of the observed effects were attributed to a direct G4 formation or to secondary effects26. Furthermore, HeLa S3 and K562 cells exposed to the G4-stabilizing molecule TMPyP4, showed genome-wide transcriptional effects. However, a smaller set of affected genes (69 and 87 genes for HeLa S3 and K562 cells, respectively) were identified at prolonged (48 h) time of exposure to relatively high TMPyP4 concentration27,28 (100 μM).
In this study, upon cell treatment with AQ1, we noticed a dose-dependent increasing number of DEGs; moreover, an overall major inhibitory transcriptional effect was observed in both cell lines. This is in line with several studies indicating G4 as a transcription suppressor element41,42,43. The ligand-induced stabilization of G4 motifs could lead to “jamming” of the transcription bubble, thus resulting in stalling of RNA polymerase and in an aborted transcription27,44. This is also in agreement with the suppression of gene expression that we observed in our experimental conditions. The use of EuQuad clarified that most of up- or downregulated DEGs possessed at least one G3 L1-7 motif near the TSS. Nevertheless, this is just an in-silico prediction and other approaches, e.g. the Chem-seq technique and G4-Seq or G4 ChIP-seq methods, could give more precise information about AQ1 and G4 interactions on the whole genome level45,46,47.
On a comparative basis, we observed a higher number of DEGs in the human versus the dog cell line. This result supports preliminary analysis on C2 cells where a slight reduction of KIT mRNA and protein levels was observed upon treatment with AQ133. This can be reasonably connected to the different sequence and distribution of the G4 forming domains in the human versus canine promoters although the modulation of distinct cellular pathways cannot be ruled out. Indeed, a critical issue might be represented by the fact that the two cell lines we used in this study represent, in both species, suitable in vitro models for mast cell diseases. However, they derive from different neoplasms, i.e. human mastocytosis (HMC 1.2) and cutaneous mast cell tumor (C2). Hence, this study does not represent a mere transcriptome comparison between the two species since apparent contradictions might result from gene and/or pathways differential expression in the two cell lines. However, despite the possible presence of substantial transcriptional differences between the two cell lines, several common cancer hallmarks and oncogene-related pathways were identified. In particular, the mTOR- and MYC-dependent pathways were downregulated in both species, whereas the apoptosis and p53-mediated programmed cell death were positively enriched. Other ligands have already been investigated for their ability to stabilize the G4 structure of MYC and KIT promoter, thereby affecting their downstream pathways. In Goh et al. (2016), a salicylaldehyde semicarbazone copper (II) complex induced apoptosis through the caspase-dependent pathway by reducing the expression of RAS and MYC, and by inhibiting Ras-ERK and PI3K-Akt pathways in MOLT-4 leukemia cells48. In 2013, Shen and colleagues proved that a G4 ligand named SYUIQ-FM05 suppressed KIT mRNA transcription and total kit protein amount in K562 cells, and this inhibition led to a downregulation of MEK activity, ERK phosphorylation, and the RAS/MEK/ERK signaling pathway49.
If we consider as a whole the GSEA hallmarks and KEGG pathways upregulated by AQ1, there is a predominance of immune system-related pathways. In this respect, no information is available on the stimulation of immune system by other G4 ligands; hence, this could represent an interesting and positive aspect to be considered in further studies.
Assuming a major effect of AQ1 at oncogenes promoters possessing G4 structures, we analyzed the most important pathways regulated by some of the most common oncogenes, such as MYC, BCL2, PDGF, hTERT and VEGF. Considering MYC-related pathway, AQ1 caused the downregulation of E2F, MDM2, p53, p73 and BCL6 in HMC1.2 cells. Interestingly, the latter two genes showed a similar behavior also in canine cells. BCL2 was modulated by the AQ1 treatment in both cell lines with the consequent increase in apoptosis-related pathways confirmed by the increasing expression of pro-apoptotic factors as BIK, BMF and NOXA (the last one only in HMC1.2 cells). In general, it is well demonstrated as anthraquinone derivatives have been shown to induce apoptosis in vitro, and such a phenomenon usually implies a decrease of BCL2 mRNA/protein50,51,52,53. The PDGF signaling pathway was affected only in HMC1.2 cells, with the downregulation of TIAM2, VAV1, VAV2 and NF-kβ. No variation in mRNA levels of TERT gene were noticed in both cell lines, despite the important role of G4 sequences in its regulation and transcription54. Finally, VEGF pathway was only partially influenced by AQ1 in human cells, as shown by caspase 9, GRB2 and SOS downregulation. In dog cell line, VEGF pathway was negatively enriched by GSEA, likewise to human.
Interestingly, in this study we identified new oncogenes affected by AQ1; for example, the gene FYN was downregulated in both cell lines. FYN is a tyrosine kinase proto-oncogene with a role in proliferation, invasion and migration in human thyroid carcinoma55,56. It also plays a critical role in the development, progression and resistance to anti-cancer drugs in breast cancer, prostate cancer and leukemia56. No information is available about the presence of G4 in its promoter sequence. On the other hand, two G4 forming sites are present in the human EGFR promoter57, a transmembrane protein activated by the epidermal growth factor and the transforming growth factor alpha (TGFα). This oncogene was downregulated by AQ1 only in dog, but other G4 ligands have been shown to affect its mRNA levels in a previous human transcriptomic study26.
The proto-oncogene JUN was upregulated in HMC1.2 (FC = 18) but not in canine cells. Consistent knowledge shows that c-jun contributes to tumor initiation and increased invasiveness58. However, few studies discovered some alternative activities of c-jun, suggesting that it may act as a double-edge sword in cancer58. As an example, c-jun seems to prevent methylation silencing of p16INK4a, a tumor suppressor and a cell cycle inhibitor gene59. Moreover, the anticancer mechanism of action of tylophorine, a plant-derived alkaloid, is mediated by c-jun, resulting in G1 cell cycle arrest in carcinoma cells, occurring through the downregulation of cyclin A260.
Despite the innovative results obtained in this study, several limitations should be considered. Probably, the most important one is the absence of a validation step for the RNA-Seq platform at the protein level. The post-transcriptional mechanisms of regulation and the emerging evidence that 5′- and 3′-untranslated regions (5′- and 3′-UTRs) as well as open reading frames (ORFs) contain putative RNA G4, might play an important role in tumor biology, e.g., switching on/off some tumor-related pathways61. From previous data, the AQ1-mediated downregulation of KIT was confirmed also at the protein level in human and canine in vitro models31,32,33. Hence, the protein evaluation of the kit-downstream pathways could substantiate current transcriptional results. Secondly, in the present study we considered one G4 ligand compound and only tumor cell lines. In perspective, it could be of interest to extend the RNA-Seq analysis also to a non-tumor cell line, to distinguish any non-specific effects. From previous information obtained in MDCK cells, these are much more resistant to AQ1 in term of cytotoxicity and survival33, but a more in-depth study investigating also non tumor-specific pathways is still missing. Finally, in terms of pathways analysis (DAVID), a limit derived by the actual scarce annotation of dog genome in comparison with the human one, and this should be consistently taken into consideration in the approach to future studies. Nevertheless, the present work represents the first step to validate dog as suitable translational model with reference to the use of G4-binding compounds.
In conclusion, the present study showed that AQ1 G4-binding compound inhibited KIT expression and its downstream signaling molecules GRB2, AKT and FYN in two in vitro models of human and canine mast cell neoplasms (HMC1.2 and C2 cells). Interestingly, besides KIT, AQ1 negatively affected the MYC-related pathway whereas it induced both apoptosis and P53-related pathways. Overall, these results suggest a possible role of AQ1 in blocking mast cells proliferation via different pathways, thus representing a potential therapeutic target for comparative mast cell tumors.
Methods
All methods were carried out in accordance with relevant guidelines and regulations generally used for handling human and animal cell lines in the Department of Comparative Biomedicine and Food Science and at the University of Padua.
Ligand
AQ1 was synthesized by Prof. G. Zagotto (University of Padua, Italy), and its chemical structure and affinity studies with h_kit1 and h_kit2 G4 sequences were previously published31. Stock solutions (10 mM) of the ligand were prepared in DMSO (Sigma-Aldrich Co., St. Louis, USA).
Cell culture
The human mast cell leukemia HMC1.2 expressing mutated KIT (D816V substitution) was kindly provided by Dr. Joseph Butterfield (Mayo Clinic, Rochester, MN, USA). The canine MCT cell line C2, expressing mutated KIT (48 bp internal tandem duplication in the juxtamembrane domain) was kindly provided by Dr. Patrice Dubreuil (Centre de Recherche en Cancérologie de Marseille, France). Cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS), 2 mM L-glutamine, 1% penicillin/streptomycin (Gibco, Thermo Scientific, Waltham, USA). The C2 culture medium was supplemented with 1 mM sodium pyruvate.
Cell viability was checked by using the Trypan Blue dye exclusion test (Sigma-Aldrich Co., St. Louis, USA). For all the experiments, cells were used from passage 5 to passage 30. Cell cultures were screened routinely for Mycoplasma spp. contamination through PCR Mycoplasma Test Kit (PromoKine, Heidelberg, Germany).
For the dose-response assay, 2 × 104 cells were seeded in P96 well plates and treated with a range of doses comprised between 0.2 µM and 10 µM (final concentrations). Cell proliferation was assessed by CellTiter-Blue® Cell Viability Assay (Alamar Blue, Promega, Madison, USA) at T12. Fluorescence was measured at 560 nm (excitation wavelength) and 590 nm (emission wavelength), by using a VICTOR™X4 Multilabel Plate Reader (Perkin Elmer, Waltham, USA). Three independent experiments were performed, and each concentration was tested six times.
Cell treatment and qPCR
Cells were seeded in 6-well plates (9 × 105 cells/well) and treated with two doses of AQ1 (1 µM and 2 µM) for 12 hours. For each experiment, we included also untreated and DMSO-treated cells. Five different experiments were conducted for each cell line.
Total RNA was extracted by RNeasy Mini Kit (Qiagen Italia, Milano, Italy). To reduce the possible presence of genomic DNA contamination, a 15-minutes on-column DNase digestion step was included in the RNA isolation protocol. Total RNA concentration was determined using the NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies Inc., Wilmington, USA), and its quality was assessed using the 2100 Bioanalyzer and RNA 6000 Nano kit (Agilent Technologies, Santa Clara, CA, USA). The reverse-transcription of mRNA into cDNA were carried out as previously published31.
The list of primers used in the present study for qPCR analysis is reported in Supplementary Table S6.
Target genes to be tested in qPCR cross-validation assay were selected considering different expression trends in both cell lines. Specifically, we selected two genes that were downregulated (RPTOR and HDAC4) as well as two other ones that were shown to be upregulated (BMF and CDKN1A) in both cell lines. We also included two DEGs that were downregulated (EGFR and KRS2) and two genes upregulated (TNFRSF11b and FAXDC2) exclusively in C2 cell line; finally, two DEGs that were downregulated (CDK6 and MID1) and two genes upregulated (EGR2 and CYP1A1) exclusively in HMC1.2 cell line. Oligonucleotide primers were designed using the software UPL Assay Design Centre web service (Roche Diagnostics, Mannheim, Germany), and each assay specificity was evaluated in silico, by using the BLAST tool.
Quantitative real-time PCR reactions (10 µL final volume) were performed as previously reported28, using 0.83 ng −1 ng of cDNA and 1–1.25 ng for C2 and HMC1.2, respectively. The analysis was conducted in a LightCycler 480 Instrument (Roche Applied Science, Indianapolis, IN) using standard qPCR conditions (95 °C for 10 min; 45 cycles at 95 °C for 10 s and at 60 °C for 30 s; 40 °C for 30 s). Calibration curves were performed using serial dilutions of a cDNA pool and corresponding values of slope, efficiency (E) and dynamic range are reported in Supplementary Table S7. The assays with an E (%) between 90% and 110% were considered efficient.
The obtained qPCR data were analyzed using the LightCycler480 software release 1.5.0 (Roche Applied Science, Indianapolis, USA) and the second derivative method; the relative quantification (RQ) was calculated with the ΔΔCt method62. For the normalization step, four internal control genes (ICGs) were used: for C2, the transmembrane BAX inhibitor motif containing 4 (CGI-119) and the CCZ1 vacuolar protein trafficking and biogenesis associated homolog (CCZ1); for HMC1.2, the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and the beta-2-microglobulin (ß2M). A cDNA pool was used as calibrator.
Library preparation and RNA-Seq analysis
Strand-specific RNA-Seq libraries were prepared using the SureSelect strand-specific mRNA library preparation kit (Agilent Technologies, Santa Clara, CA, USA) as per the manufacturer’s instructions. In brief, poly(A) RNA was purified from 1 µg of total RNA using two serial rounds of binding to oligo(dT) magnetic particles; then, fragmented RNA was reverse transcribed to generate cDNA. An Illumina-specific adaptor was sequentially ligated to the 3′ end of cDNA fragments, purified using the AMPure XP beads (Beckman Coulter, Brea, CA, USA), and finally PCR-amplified (13 cycles) using an appropriate indexing primer to allow further samples multiplexing. The PCR-amplified libraries were purified with the AMPure XP beads (Beckman Coulter, Brea, CA, USA) and, then, assessed for their quality and fragments distribution using the 2100 Bioanalyzer DNA 1000 assay (Agilent Technologies, Santa Clara, CA, USA). In the presence of adaptor-dimers (Electropherogram’s peak at 100 to 150-bp), another round of magnetic beads purification was performed. Libraries were quantified using both the Qubit® Fluorometer (Life Technologies, USA) and the qPCR-based NEBNext library quantification kit (New England BioLabs, UK). Finally, equimolar amounts of each ten index-tagged libraries were multiplexed together in one pool (total 4 pools) and then sequenced by an Illumina HiSeq4000 for 50 sequencing cycles. The raw 50 bp single-end sequences (Sanger/Illumina 1.9 encoding) were quality-controlled using FastQC v.0.11.4 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/), the low-quality bases (quality scores <30) and the adaptor contamination (if present) were removed by Trimmomatic v.0.3663 using the parameters ‘LEADING:3 SLIDINGWINDOW:4:20 MINLEN:25′. In the quality control step, we eliminated one biological replicate that showed a non-sufficient output. The high-quality reads were mapped by HISAT2 v.2.0.564 against the Ensembl reference genomes of Canis lupus familiaris (CanFam3.1, ftp://ftp.ensembl.org/pub/release-90/fasta/canis_familiaris/dna) or Homo sapiens (GRCh38, ftp://ftp.ensembl.org/pub/release-90/fasta/homo_sapiens/dna/). The uniquely-mapped reads aligned to exons were counted with HTSeq v.0.6.1, then tested using the DESeq2 R package v.1.14.1 for the presence of DEGs65,66. Genes with a FDR less than 0.05 and a FC value more than 2 were considered as DEGs. The sequencing data associated with this project were deposited in the GenBank’s Gene Expression Omnibus (GEO) under the accession number GSE120272. The statistical analysis of RNA-Seq data was performed considering AQ1 (1 or 2 µM) versus DMSO in each cell line. In order to establish the congruency among biological replicates, a PCA and a hierarchical clustering were performed.
References
Mashima, T. et al. Comprehensive transcriptomic analysis of molecularly targeted drugs in cancer for target pathway evaluation. Cancer Sci. 106, 909–920 (2015).
LeBlanc, A. K., Mazcko, C. & Khanna, C. Defining the value of a comparative approach to cancer drug development. Clin. Cancer Res. 22, 2133–2138 (2016).
Abou-Jawde, R., Choueiri, T., Alemany, C. & Mekhail, T. An overview of targeted treatments in cancer. Clin. Ther. 25, 2121–2137 (2003).
Dubreuil, P. et al. Masitinib (AB1010), a potent and selective tyrosine kinase inhibitor targeting KIT. Plos One 4, e7258 (2009).
Deininger, M. W. N. & Druker, B. J. Specific targeted therapy of chronic myelogenous leukemia with imatinib. Pharmacol. Rev. 55, 401–423 (2003).
Rivera, P. et al. Mammary tumor development in dogs is associated with BRCA1 and BRCA2. Cancer Res. 69, 8770–8774 (2009).
Fowles, J. S., Brown, K. C., Hess, A. M., Duval, D. L. & Gustafson, D. L. Intra-and interspecies gene expression models for predicting drug response in canine osteosarcoma. BMC Bioinform. 17, 93–107 (2016).
Clemente-Vicario, F. et al. Human Genetic Relevance and Potent Antitumor Activity of Heat Shock Protein 90 Inhibition in Canine Lung Adenocarcinoma Cell Lines. PLoS One 10, e0142007 (2015).
Dhawan, D. et al. Comparative gene expression analyses identify luminal and basal subtypes of canine invasive urothelial carcinoma that mimic patterns in human invasive bladder cancer. PLoS One 10, e0136688 (2015).
Ramsey, S. A. et al. Cross-species analysis of the canine and human bladder cancer transcriptome and exome. Genes Chromosom. Cancer 56, 328–343 (2017).
Richards, K. L. et al. Gene profiling of canine B-cell lymphoma reveals germinal center and postgerminal center subtypes with different survival times, modeling human DLBCL. Cancer Res. 73, 5029–39 (2013).
Schiffman, J. D. & Breen, M. Comparative oncology: what dogs and other species can teach us about humans with cancer. Philos Trans R Soc Lond B Biol Sci. 370, 20140231 (2015).
Fowles, J. S., Denton, C. L. & Gustafson, D. L. Comparative analysis of MAPK and PI3K/AKT pathway activation and inhibition in human and canine melanoma. Vet. Comp. Oncol. 13, 288–304 (2013).
Kumar, N. & Maiti, S. A thermodynamic overview of naturally occurring intramolecular DNA quadruplexes. Nucleic Acids Res. 36, 5610–5622 (2008).
Wu, Y. & Brosh, R. M. G-quadruplex nucleic acids and human disease. FEBS J. 277, 3470–3488 (2010).
Agrawal, P., Lin, C., Mathad, R. I., Carver, M. & Yang, D. The Major G-Quadruplex Formed in the Human BCL-2 Proximal Promoter Adopts a Parallel Structure with a 13-nt Loop in K+ Solution. J. Am. Chem. Soc. 136, 1750–1753 (2014).
Du, Z., Zhao, Y. & Li, N. Genome-wide analysis reveals regulatory role of G4 DNA in gene transcription. Genome Res. 18, 233–241 (2008).
Siddiqui-Jain, A., Grand, C. L., Bearss, D. J. & Hurley, L. H. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. USA 99, 11593–11598 (2002).
Gunaratnam, M. et al. Targeting human gastrointestinal stromal tumor cells with a quadruplex-binding small molecule. J. Med. Chem. 52, 3774–3783 (2009).
Sun, D. et al. The proximal promoter region of the human vascular endothelial growth factor gene has a G-quadruplex structure that can be targeted by G-quadruplex-interactive agents. Mol. Cancer. Ther. 7, 880–889 (2008).
Chen, Y. et al. The Major G-Quadruplex Formed in the Human Platelet-Derived Growth Factor Receptor β (PDGFR-β) Promoter Adopts a Novel Broken-Strand Structure in K+ Solution. J Am Chem Soc. 134, 13220–13223 (2012).
Welsh, S. J. et al. Inhibition of the hypoxia-inducible factor pathway by a G-quadruplex binding small molecule. Sci Rep. 3, 2799–2808 (2013).
Rankin, S. et al. Putative DNA quadruplex formation within the human c-kit oncogene. J Am Chem Soc. 127, 10584–10589 (2005).
Fernando, H. et al. A conserved quadruplex motif located in a transcription activation site of the human c-kit oncogene. Biochemistry 45, 7854–7860 (2006).
Agarwal, T., Roy, S., Kumar, S., Chakraborty, T. K. & Maiti, S. In the sense of transcription regulation by G-Quadruplexes: asymmetric effects in sense and antisense strands. Biochem. 53, 3711–3718 (2014).
Halder, R., Riou, J. F., Teulade-Fichou, M. P., Frickey, T. & Hartig, J. S. Bisquinolinium compounds induce quadruplex specific transcriptome changes in HeLa S3 cell lines. BMC Res. Notes 5, 138–149 (2012).
Verma, A. et al. Genome-Wide Computational and Expression Analyses Reveal G-Quadruplex DNA Motifs as Conserved cis-Regulatory Elements in Human and Related Species. J. Med. Chem. 51, 5641–5649 (2008).
Mikami-Terao, Y. et al. Antitumor activity of G-quadruplex-interactive agent TMPyP4 in K562 leukemic cells. Cancer Lett. 261, 226–234 (2008).
Zheng, X. et al. TMPyP4 promotes cancer cell migration at low doses, but induces cell death at high doses. Sci. Rep. 6, 26592, https://doi.org/10.1038/srep26592 (2016).
Marchetti, C. et al. Targeting Multiple Effector Pathways in Pancreatic Ductal Adenocarcinoma with a G-Quadruplex-Binding Small Molecule. J Med Chem. 61, 2500–2517 (2018).
Zorzan, E. et al. Screening of candidate G-quadruplex ligands for the human c-KIT promotorial region and their effects in multiple in-vitro models. Oncotarget 7, 21658–21675 (2016).
Da Ros, S. et al. Sequencing and G-quadruplex folding of the canine proto-oncogene KIT promoter region: might dog be used as a model for human disease? Plos One 9, 1–11 (2014).
Zorzan, E. et al. Targeting canine KIT promoter by candidate DNA G-quadruplex ligands. J. Pharmacol. Exp. Ther. https://doi.org/10.1124/jpet.118.248997 (2018).
Kanehisa, M. et al. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 45, D353–D361 (2017).
Kanehisa, M. et al. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44, D457–D462 (2016).
Kanehisa, M. & Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28, 27–30 (2000).
Liang, J. et al. The C-Kit Receptor-Mediated Signal Transduction and Tumor-Related Diseases. Int. J. Biol. Sci. 9, 435–443 (2013).
Yadav, V. K., Abraham, J. K., Mani, P., Kulshrestha, R. & Chowdhury, S. QuadBase: genome-wide database of G4 DNA occurrence and conservation in human, chimpanzee, mouse and rat promoters and 146 microbes. Nucleic Acids Res. 36, D381–385 (2008).
Dhapola, P. & Chowdhury, S. QuadBase2: web server for multiplexed guanine quadruplex mining and visualization. Nucleic Acids Res. 44, W277–W283 (2016).
Ilyinsky, N. S. et al. Quadruplex Ligands: Mechanisms of Anticancer Action and Target Binding. Mol Biol 48, 778–794 (2014).
Balasubramanian, S., Hurley, L. H. & Neidle, S. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy? Nat. Rev. Drug Discov. 10, 261–275 (2011).
Wang, X. D. et al. Turning off transcription of the bcl-2 gene by stabilizing the bcl-2 promoter quadruplex with quindoline derivatives. J. Med. Chem. 53, 4390–4398 (2010).
Eddy, J. & Maizels, N. Selection for the G4 DNA motif at the 5′end of human genes. Mol. Carcinog. 48, 319–325 (2009).
Tateishi-Karimata, H. et al. Destabilization of DNA G-Quadruplexes by Chemical Environment Changes during Tumor Progression Facilitates Transcription. J Am Chem Soc. 140, 642–651 (2018).
Rodriguez, R. & Miller, K. M. Unravelling the genomic targets of small molecules using high-throughput sequencing. Nat Rev Genet. 15, 783–796 (2014).
Chambers, V. S. et al. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat Biotechnol. 33, 877–881 (2015).
Hänsel-Hertsch, R. et al. Genome-wide mapping of endogenous G-quadruplex DNA structures by chromatin immunoprecipitation and high-throughput sequencing. Nat Protoc. 13, 551–564 (2018).
Goh, Y. et al. Downregulation of oncogenic RAS and c-Myc expression in MOLT-4 leukemia cells by a salicylaldehyde semicarbazone copper(II) complex. Sci Rep. 6, 36868, https://doi.org/10.1038/srep36868 (2016).
Shen, F. H. et al. The G-quadruplex ligand, SYUIQ-FM05, targets proto-oncogene c-kit transcription and induces apoptosis in K562 cells. Pharm Biol. 51, 447–454 (2013).
Dong, X. et al. Induction of apoptosis in HepaRG cell line by aloe-emodin through generation of reactive oxygen species and the mitochondrial pathway. Cell. Physiol. Biochem. 42, 685–696 (2017).
Hasan, T. N., B, L. G., Shafi, G., Al-Hazzani, A. A. & Alshatwi, A. A. Anti-proliferative effects of organic extracts from root bark of Juglans regia L. (RBJR) on MDA-MB-231 human breast cancer cells: role of Bcla-2/Bax, caspases and Tp53. Asian Pac. J. Cancer Prev. 12, 525–30 (2011).
Huang, L. et al. Anthraquinone G503 induces apoptosis in gastric cancer cells through the mitochondrial pathway. Plos One 9, e108286 (2014).
Huang, Q., Lu, G., Shen, H., Chung, M. C. M. & Ong, C. N. Anti-cancer properties of anthraquinones from rhubarb. Med Res Rev 27, 609–630 (2007).
Gomez., D. et al. Telomerase downregulation induced by the G-quadruplex ligand 12459 in A549 cells is mediated by hTERT RNA alternative splicing. Nucleic Acids Res. 32, 371–379 (2004).
Elias, D. & Ditzel, H. J. Fyn is an important molecule in cancer pathogenesis and drug resistance. Pharmacol Res. 100, 250–254 (2015).
Jing, Z., Huiling, L., Dongting, X. & Huijuan, Z. Upregulation of Tyrosine Kinase FYN in Human Thyroid Carcinoma: Role in Modulating Tumor Cell Proliferation, Invasion, and Migration. Cancer Biother. Radiopharm. 32, 320–326 (2017).
Greco, M. L. et al. Coexistence of two main folded G-quadruplexes within a single G-rich domain in the EGFR promoter. Nucleic Acids Res. 45, 10132–10142 (2017).
Shaulian, E. AP-1 The Jun proteins: Oncogenes or tumor suppressors in disguise? Cell. Sign. 22, 894–899 (2010).
Kollmann, K., Heller, G. & Sexl, V. c-JUN prevents methylation of p16(INK4a) (and Cdk6): the villain turned bodyguard. Oncotarget 2, 422–427 (2011).
Yang, C. W. et al. c-Jun-mediated anticancer mechanisms of tylophorine. Carcinogenesis 34, 1304–1314 (2013).
Song, J., Perreault, J. P., Topisirovic, I. & Richard, S. RNA G-quadruplexes and their potential regulatory roles in translation. Translation 4, e1244031 (2016).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408 (2001).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast-spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015).
Anders, S., Pyl, P. T. & Huber, W. HTSeq - a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome biol. 15, 550–571 (2014).
Acknowledgements
This project was supported by grants from the University of Padua (prot. CPDA147272 and DOR1798331) to C.S., M.G. and M.D. respectively. E.Z. and R.E. were recipient of a one year post-doc grant fellowship from the University of Padua. Authors kindly acknowledge Kanehisa Laboratories for the permission of using KEGG results and images (ref. 180271).
Author information
Authors and Affiliations
Contributions
E.Z., M.G., M.D. and C.S. designed the study. E.Z. and R.E. performed the experiments and analyzed results. E.Z. and R.E. wrote the paper. All authors reviewed the article and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zorzan, E., Elgendy, R., Giantin, M. et al. Whole-Transcriptome Profiling of Canine and Human in Vitro Models Exposed to a G-Quadruplex Binding Small Molecule. Sci Rep 8, 17107 (2018). https://doi.org/10.1038/s41598-018-35516-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-35516-y
Keywords
This article is cited by
-
Bioactive nutraceuticals as G4 stabilizers: potential cancer prevention and therapy—a critical review
Naunyn-Schmiedeberg's Archives of Pharmacology (2023)
-
Ligands stimulating antitumour immunity as the next G-quadruplex challenge
Molecular Cancer (2022)
-
Multiple G-quadruplex binding ligand induced transcriptomic map of cancer cell lines
Journal of Cell Communication and Signaling (2022)
-
G-quadruplexes: a promising target for cancer therapy
Molecular Cancer (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.