Ensoma is poised to create a new therapeutic category. Leveraging first-in-class delivery and engineering, the biotech aims to harness the power of hematopoietic stem cells (HSCs) in vivo to provide one-time, off-the-shelf treatments for cancer, autoimmunity, and genetic diseases.
Using its virus-like particle (VLP) delivery platform, Ensoma is focused on treating disease through engineering of blood and immune cells inside the body. The potential to treat more diseases with gene-editing approaches led Ensoma to acquire Twelve Bio in early 2023, expanding its engineering toolkit to include clustered regularly interspaced short palindromic repeats (CRISPR)–associated protein 12a (Cas12a)-based editors.
The company has integrated these editors into its in vivo Engenious platform and pipeline programs, and is exploring opportunities to partner its editors with companies targeting other cells using a variety of delivery technologies.
Ensoma’s toolkit is built on structural insights. Using X-ray crystallography and cryogenic electron microscopy, Twelve Bio’s University of Copenhagen founders showed that Cas12a recognizes DNA target sequences with exquisite specificity; studied molecular interactions between the protein, targeting CRISPR RNA (crRNA), and target sequence DNA; and revealed how the enzyme changes shape to accommodate precise binding.
The insights equipped the company to enhance the natural advantages of Cas12a, a small, precise editing protein, to create editors with improved safety potential, better multiplex capabilities, and delivery platform versatility (Fig. 1).
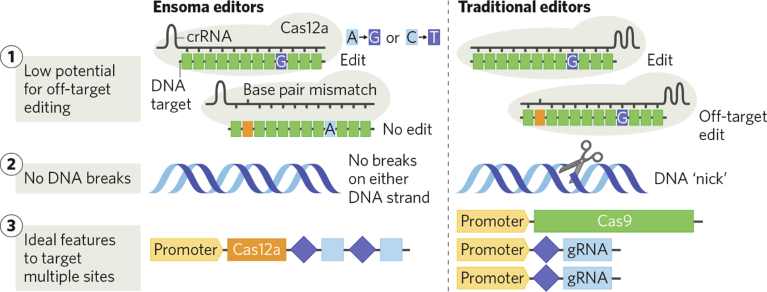
Fig. 1 | Ensoma’s editors. The high-efficiency base editors have low off-target potential, no DNA breaks, and improved multiplex capabilities compared with traditional editors. Cas, CRISPR-associated protein; CRISPR, clustered regularly interspaced short palindromic repeats; crRNA, CRISPR RNA; gRNA, guide RNA.
High-specificity editors without DNA breaks
CRISPR–Cas12a has a low tolerance for base-pair mismatches between the crRNA and the DNA target sequence. Ensoma applied its knowledge of the protein’s structure to further hone specificity and achieve safer in vivo-editing potential. The company studied enzyme specificity in collaboration with Shengdar Tsai, a world-leading expert in genome-editing technology and off-target analysis, at St. Jude Children’s Research Hospital.
How Ensoma edits DNA is another differentiator. First-generation CRISPR systems cause double-strand breaks that may lead to disease-causing chromosomal rearrangements1. Alternative, ‘gentler’ editing approaches, such as base and prime editing, only cut one strand of DNA, but a recent publication showed that these nicks can also cause rearrangements2. Ensoma’s editors do not cause any DNA breaks.
“Rearrangements are rare but they can induce serious diseases, even years after treatment. Since we are developing in vivo medicines, our goal was to design editors that would eliminate even potential risks and edit only the target DNA without disturbing any other part of the genome,” said Stefano Stella, VP, Gene Editing at Ensoma.
Ensoma developed its proprietary base editors to have unique capabilities that are well-suited to safe, one-time, in vivo engineering. Features such as base editing without DNA breaks set Ensoma’s platform apart from other editing technologies.
Making multiplex modifications
Although single-gene disorders will be many of the first applications for precise gene-editing technologies, treating other indications will require multiple modifications.
Ensoma’s Cas12a-based editors, unlike CRISPR-associated protein 9 (Cas9)-based systems, have specific RNA-processing activity, allowing them to process a single long RNA containing several crRNAs, which in turn enables the protein to target multiple specific DNA sequences.
Multiplex editing can also raise the intrinsic risks of gene-editing approaches that rely on DNA breaks. “Highly precise base conversions, the absence of DNA breaks, and unique RNA-processing activity make Ensoma’s base editors an elegant and potentially safer editing system for indications that may benefit from multiple modifications in vivo,” said Robert Peters, CSO at Ensoma.
Versatile editors for AAV or RNA delivery
To support RNA-based delivery, Ensoma engineered the RNA-processing activity to produce mature messenger RNA (mRNA) encoding for both the base editor and one or multiple crRNAs. This feature allows Ensoma’s editors to be deliverable using a single RNA encoding the enzyme and the crRNA(s), rather than requiring delivery of each component independently, as with Cas9-based systems.
Although Ensoma’s VLP delivery technology can accommodate very large cargo, other platforms, such as adeno-associated viruses (AAVs), have limited capacity that prevents delivery of traditional editing systems within one vector.
In contrast, Ensoma’s editors can be delivered using a single AAV capsid. Cas12a is a compact protein and, because of its specific RNA activity, can be delivered as a single gene encoding all components of the editing system. Ensoma is working to further reduce the protein’s size to unlock the full potential of delivering base editors in a single AAV.
Partnering the platform
Ensoma has built a gene-editing platform with the precision, efficiency, and compact size needed to enable programs across a wide range of tissues and delivery vehicles. The company is open to exploring partnerships to realize the full promise of its technology.
Going forward, Ensoma plans to drive its development candidates toward the clinic while continuing to advance its platform, positioning partners to benefit from enhancements—with the ultimate goal of creating one-time in vivo genomic medicines.


