Abstract
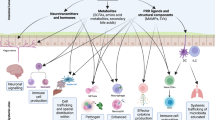
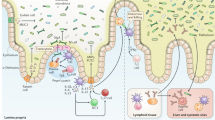
Gastrointestinal commensal microbiota is a concentrated mix of microbial life forms, including bacteria, fungi, archaea and viruses. These life forms are targets of host antimicrobial defense in order to establish a homeostatic symbiosis inside the host. However, they are also instrumental in shaping the functions of our immune system via a diverse set of communication mechanisms. In the gut, T helper 17, regulatory T and B cells are continuously tuned by specific microbial strains and metabolic processes. These cells in return help to establish a mutually beneficial exchange with the gut microbial contents. Imbalances in this symbiosis lead to dysregulations in the host’s ability to control infections and the development of autoimmune diseases. In addition, the commensal microbiota has a significant and obligatory role in shaping both gut intrinsic and distal lymphoid organs, casting a large impact on the overall immune landscape in the host. This review discusses the major components of the microbial community in the gut and how its members collectively and individually exert regulatory roles in the host immune system and lymphoid structure development, as well as the functions of several major immune cell types.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Relman DA . 'Til death do us part': coming to terms with symbiotic relationships - Foreword. Nat Rev Microbiol 2008; 6: 721–724.
Adlerberth I, Wold AE . Establishment of the gut microbiota in Western infants. Acta Paediatr 2009; 98: 229–238.
Tremaroli V, Backhed F . Functional interactions between the gut microbiota and host metabolism. Nature 2012; 489: 242–249.
Ramakrishna BS . Role of the gut microbiota in human nutrition and metabolism. J Gastroenterol Hepatol 2013; 28: 9–17.
Backhed F, Manchester JK, Semenkovich CF, Gordon JI . Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc Natl Acad Sci USA 2007; 104: 979–984.
Cash HL, Hooper LV . Commensal bacteria shape Intestinal immune system development. ASM News 2005; 71: 77–83.
Backhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA 2004; 101: 15718–15723.
Shan M, Gentile M, Yeiser JR, Walland AC, Bornstein VU, Chen K et al. Mucus enhances gut homeostasis and oral tolerance by delivering immunoregulatory signals. Science 2013; 342: 447–453.
Macpherson AJ, Uhr T . Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science 2004; 303: 1662–1665.
Palm NW, de Zoete MR, Cullen TW, Barry NA, Stefanowski J, Hao LM et al. Immunoglobulin A Coating Identifies Colitogenic Bacteria in Inflammatory Bowel Disease. Cell 2014; 158: 1000–1010.
Larsson E, Tremaroli V, Lee YS, Koren O, Nookaew I, Fricker A et al. Analysis of gut microbial regulation of host gene expression along the length of the gut and regulation of gut microbial ecology through MyD88. Gut 2012; 61: 1124–1131.
Bevins CL, Salzman NH . Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat Rev Microbiol 2011; 9: 356–368.
Gordon HA, Bruckner-Kardoss E, Wostmann BS . Aging in germ-free mice: life tables and lesions observed at natural death. J Gerontol 1966; 21: 380–387.
Bauer H, Horowitz RE, Levenson SM, Popper H . The response of the lymphatic tissue to the microbial flora. Studies on germfree mice. Am J Pathol 1963; 42: 471–483.
Ley RE, Peterson DA, Gordon JI . Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 2006; 124: 837–848.
McFall-Ngai M . Adaptive immunity: care for the community. Nature 2007; 445: 153.
Sevelsted A, Stokholm J, Bonnelykke K, Bisgaard H . Cesarean section and chronic immune disorders. Pediatrics 2015; 135: e92–e98.
Bager P, Wohlfahrt J, Westergaard T . Caesarean delivery and risk of atopy and allergic disease: meta-analyses. Clin Exp Allergy 2008; 38: 634–642.
Bauer E, Williams BA, Smidt H, Verstegen MW, Mosenthin R . Influence of the gastrointestinal microbiota on development of the immune system in young animals. Curr Issues Intest Microbiol 2006; 7: 35–51.
Berer K, Mues M, Koutrolos M, Rasbi ZA, Boziki M, Johner C et al. Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature 2011; 479: 538–541.
Adlerberth I, Wold AE . Establishment of the gut microbiota in Western infants. Acta paediatrica 2009; 98: 229–238.
Qin JJ, Li RQ, Raes J, Arumugam M, Burgdorf KS, Manichanh C et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010; 464: 59–U70.
Browne HP, Forster SC, Anonye BO, Kumar N, Neville BA, Stares MD et al. Culturing of 'unculturable' human microbiota reveals novel taxa and extensive sporulation. Nature 2016; 533: 543-+.
Abt MC, Osborne LC, Monticelli LA, Doering TA, Alenghat T, Sonnenberg GF et al. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity 2012; 37: 158–170.
Ichinohe T, Pang IK . Kumamoto Y, Peaper DR, Ho JH, Murray TS, et al. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc Natl Acad Sci USA 2011; 108: 5354–5359.
Khan KJ, Ullman TA, Ford AC, Abreu MT, Abadir A, Marshall JK et al. Antibiotic Therapy in Inflammatory Bowel Disease: A Systematic Review and Meta-Analysis. Am J Gastroenterol 2011; 106: 661–673.
Kawada M, Arihiro A, Mizoguchi E . Insights from advances in research of chemically induced experimental models of human inflammatory bowel disease. World J Gastroenterol 2007; 13: 5581–5593.
Lepage P, Hasler R, Spehlmann ME, Rehman A, Zvirbliene A, Begun A et al. Twin Study Indicates Loss of Interaction Between Microbiota and Mucosa of Patients With Ulcerative Colitis. Gastroenterology 2011; 141: 227–236.
Frank DN, Amand ALS, Feldman RA, Boedeker EC, Harpaz N, Pace NR . Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA 2007; 104: 13780–13785.
Manichanh C, Borruel N, Casellas F, Guarner F . The gut microbiota in IBD. Nat Rev Gastroenterol Hepatol 2012; 9: 599–608.
West CE . Gut microbiota and allergic disease: new findings. Curr Opin Clin Nutr Metab Care 2014; 17: 261–266.
Yokote H, Miyake S, Croxford JL, Oki S, Mizusawa H, Yamamura T . NKT cell-dependent amelioration of a mouse model of multiple sclerosis by altering gut flora. Am J Pathol 2008; 173: 1714–1723.
Lee YK, Menezes JS, Umesaki Y, Mazmanian SK . Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc Natl Acad Sci USA 2011; 108: 4615–4622.
Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 2011; 331: 337–341.
Miyake S, Kim S, Suda W, Oshima K, Nakamura M, Matsuoka T et al. Dysbiosis in the Gut Microbiota of Patients with Multiple Sclerosis, with a Striking Depletion of Species Belonging to Clostridia XIVa and IV Clusters. PLoS ONE 2015; 10: e0137429.
Ochoa-Reparaz J, Mielcarz DW, Ditrio LE, Burroughs AR, Begum-Haque S, Dasgupta S et al. Central nervous system demyelinating disease protection by the human commensal Bacteroides fragilis depends on polysaccharide A expression. J Immunol 2010; 185: 4101–4108.
Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity 2010; 32: 815–827.
Kriegel MA, Sefik E, Hill JA, Wu HJ, Benoist C, Mathis D . Naturally transmitted segmented filamentous bacteria segregate with diabetes protection in nonobese diabetic mice. Proc Natl Acad Sci USA 2011; 108: 11548–11553.
Anderson HW . Yeast-like fungi of the human intestinal tract. J Infect Dis 1917; 21: 341–U18.
Gumbo T, Isada CM, Hall G, Karafa MT, Gordon SM . Candida glabrata fungemia - Clinical features of 139 patients. Medicine (Baltimore) 1999; 78: 220–227.
Dimitrov DV . The human gutome: nutrigenomics of the host-microbiome interactions. OMICS 2011; 15: 419–430.
Rajilic-Stojanovic M, de Vos WM . The first 1000 cultured species of the human gastrointestinal microbiota. FEMS Microbiol Rev 2014; 38: 996–1047.
Underhill DM, Iliev ID . The mycobiota: interactions between commensal fungi and the host immune system. Nat Rev Immunol 2014; 14: 405–416.
Hoffmann C, Dollive S, Grunberg S, Chen J, Li H, Wu GD et al. Archaea and fungi of the human gut microbiome: correlations with diet and bacterial residents. PLoS ONE 2013; 8: e66019.
Standaert-Vitse A, Sendid B, Joossens M, Francois N, Vandewalle-El Khoury P, Branche J et al. Candida albicans colonization and ASCA in familial Crohn's disease. Am J Gastroenterol 2009; 104: 1745–1753.
Zwolinska-Wcislo M, Brzozowski T, Budak A, Kwiecien S, Sliwowski Z, Drozdowicz D et al. Effect of Candida colonization on human ulcerative colitis and the healing of inflammatory changes of the colon in the experimental model of colitis ulcerosa. J Physiol Pharmacol 2009; 60: 107–118.
Gringhuis SI, Kaptein TM, Wevers BA, Theelen B, van der Vlist M, Boekhout T et al. Dectin-1 is an extracellular pathogen sensor for the induction and processing of IL-1beta via a noncanonical caspase-8 inflammasome. Nat Immunol 2012; 13: 246–254.
LeibundGut-Landmann S, Gross O, Robinson MJ, Osorio F, Slack EC, Tsoni SV et al. Syk- and CARD9-dependent coupling of innate immunity to the induction of T helper cells that produce interleukin 17. Nat Immunol 2007; 8: 630–638.
Iliev ID, Funari VA, Taylor KD, Nguyen Q, Reyes CN, Strom SP et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science 2012; 336: 1314–1317.
Rohwer F . Global phage diversity. Cell 2003; 113: 141-.
Reyes A, Wu M, McNulty NP, Rohwer FL, Gordon JI . Gnotobiotic mouse model of phage-bacterial host dynamics in the human gut. Proc Natl Acad Sci USA 2013; 110: 20236–20241.
Ogilvie LA, Jones BV . The human gut virome: a multifaceted majority. Front Microbiol 2015; 6: 918.
Norman JM, Handley SA, Baldridge MT, Droit L, Liu CY, Keller BC et al. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell 2015; 160: 447–460.
Falk PG, Hooper LV, Midtvedt T, Gordon JI . Creating and maintaining the gastrointestinal ecosystem: what we know and need to know from gnotobiology. Microbiol Mol Biol Rev 1998; 62: 1157–1170.
Macpherson AJ, Harris NL . Interactions between commensal intestinal bacteria and the immune system. Nat Rev Immunol 2004; 4: 478–485.
Bouskra D, Brezillon C, Berard M, Werts C, Varona R, Boneca IG et al. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature 2008; 456: 507–510.
Shulzhenko N, Morgun A, Hsiao W, Battle M, Yao M, Gavrilova O et al. Crosstalk between B lymphocytes, microbiota and the intestinal epithelium governs immunity versus metabolism in the gut. Nat Med 2011; 17: 1585–U97.
Fransen F, Zagato E, Mazzini E, Fosso B, Manzari C, El Aidy S et al. BALB/c and C57BL/6 mice differ in polyreactive IgA abundance, which impacts the generation of antigen-specific IgA and microbiota diversity. Immunity 2015; 43: 527–540.
Peterson DA, McNulty NP, Guruge JL, Gordon JI . IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe 2007; 2: 328–339.
Suzuki K, Meek B, Doi Y, Muramatsu M, Chiba T, Honjo T et al. Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proc Natl Acad Sci USA 2004; 101: 1981–1986.
Planer JD, Peng Y, Kau AL, Blanton LV, Ndao IM, Tarr PI et al. Development of the gut microbiota and mucosal IgA responses in twins and gnotobiotic mice. Nature 2016; 534: 263–266.
Brandtzaeg P . Mucosal immunity: induction, dissemination, and effector functions. Scand J Immunol 2009; 70: 505–515.
Sato A, Hashiguchi M, Toda E, Iwasaki A, Hachimura S, Kaminogawa S . CD11b+ Peyer's patch dendritic cells secrete IL-6 and induce IgA secretion from naive B cells. J Immunol 2003; 171: 3684–3690.
Spits H, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G et al. Innate lymphoid cells—a proposal for uniform nomenclature. Nat Rev Immunol 2013; 13: 145–149.
Bjersing JL, Telemo E, Dahlgren U, Hanson LA . Loss of ileal IgA+ plasma cells and of CD4+ lymphocytes in ileal Peyer's patches of vitamin A deficient rats. Clin Exp Immunol 2002; 130: 404–408.
Mora JR, Iwata M, Eksteen B, Song SY, Junt T, Senman B et al. Generation of gut-homing IgA-secreting B cells by intestinal dendritic cells. Science 2006; 314: 1157–1160.
Tokuyama H, Tokuyama Y . Retinoids enhance Iga production by lipopolysaccharide-stimulated murine spleen-cells. Cell Immunol 1993; 150: 353–363.
Tokuyama H, Tokuyama Y . Retinoids enhance IgA production by lipopolysaccharide-stimulated murine spleen cells. Cell Immunol 1993; 150: 353–363.
Watanabe K, Sugai M, Nambu Y, Osato M, Hayashi T, Kawaguchi M et al. Requirement for Runx proteins in IgA class switching acting downstream of TGF-beta 1 and retinoic acid signaling. J Immunol 2010; 184: 2785–2792.
Seo GY, Jang YS, Kim HA, Lee MR, Park MH, Park SR et al. Retinoic acid, acting as a highly specific IgA isotype switch factor, cooperates with TGF-beta 1 to enhance the overall IgA response. J Leukoc Biol 2013; 94: 325–335.
Cong Y, Feng T, Fujihashi K, Schoeb TR, Elson CO . A dominant, coordinated T regulatory cell-IgA response to the intestinal microbiota. Proc Natl Acad Sci USA 2009; 106: 19256–19261.
Kawamoto S, Maruya M, Kato LM, Suda W, Atarashi K, Doi Y et al. Foxp3(+) T cells regulate immunoglobulin a selection and facilitate diversification of bacterial species responsible for immune homeostasis. Immunity 2014; 41: 152–165.
Diehl GE, Longman RS, Zhang JX, Breart B, Galan C, Cuesta A et al. Microbiota restricts trafficking of bacteria to mesenteric lymph nodes by CX(3)CR1(hi) cells. Nature 2013; 494: 116–120.
Abbas AK, Benoist C, Bluestone JA, Campbell DJ, Ghosh S, Hori S et al. Regulatory T cells: recommendations to simplify the nomenclature. Nat Immunol 2013; 14: 307–308.
Josefowicz SZ, Lu LF, Rudensky AY . Regulatory T cells: mechanisms of differentiation and function. Annu Rev Immunol 2012; 30: 531–564.
Wirnsberger G, Hinterberger M, Klein L . Regulatory T-cell differentiation versus clonal deletion of autoreactive thymocytes. Immunology and cell biology 2011; 89: 45–53.
Hsieh CS, Liang Y, Tyznik AJ, Self SG, Liggitt D, Rudensky AY . Recognition of the peripheral self by naturally arising CD25+ CD4+ T cell receptors. Immunity 2004; 21: 267–277.
Schmitt EG, Williams CB . Generation and function of induced regulatory T cells. Front Immunol 2013; 4: 152.
Sakaguchi S, Sakaguchi N, Asano M, Itoh M, Toda M . Immunologic self-tolerance maintained by activated T cells expressing IL-2 receptor alpha-chains (CD25). Breakdown of a single mechanism of self-tolerance causes various autoimmune diseases. J Immunol 1995; 155: 1151–1164.
Mottet C, Uhlig HH, Powrie F . Cutting edge: Cure of colitis by CD4(+) CD25(+) regulatory T cells. J Immunol 2003; 170: 3939–3943.
Weiss JM, Bilate AM, Gobert M, Ding Y, Curotto de Lafaille MA, Parkhurst CN et al. Neuropilin 1 is expressed on thymus-derived natural regulatory T cells, but not mucosa-generated induced Foxp3+ T reg cells. J Exp Med 2012; 209: 1723–1742, S1.
Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 2013; 500: 232–236.
Kim KS, Hong SW, Han D, Yi J, Jung J, Yang BG et al. Dietary antigens limit mucosal immunity by inducing regulatory T cells in the small intestine. Science 2016; 351: 858–863.
Coombes JL, Siddiqui KR, Arancibia-Carcamo CV, Hall J, Sun CM, Belkaid Y et al. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-beta and retinoic acid-dependent mechanism. J Exp Med 2007; 204: 1757–1764.
Lathrop SK, Bloom SM, Rao SM, Nutsch K, Lio CW, Santacruz N et al. Peripheral education of the immune system by colonic commensal microbiota. Nature 2011; 478: 250–254.
Nordlund E, Aura AM, Mattila I, Kosso T, Rouau X, Poutanen K . Formation of phenolic microbial metabolites and short-chain fatty acids from rye, wheat, and oat bran and their fractions in the metabolical in vitro colon model. J Agric Food Chem 2012; 60: 8134–8145.
Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 2013; 341: 569–573.
Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 2013; 504: 451–455.
Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013; 504: 446–450.
Singh N, Gurav A, Sivaprakasam S, Brady E, Padia R, Shi H et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 2014; 40: 128–139.
Round JL, Mazmanian SK . Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci USA 2010; 107: 12204–12209.
Mazmanian SK, Round JL, Kasper DL . A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 2008; 453: 620–625.
Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA et al. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science 2011; 332: 974–977.
Martin-Orozco N, Chung Y, Chang SH, Wang YH, Dong C . Th17 cells promote pancreatic inflammation but only induce diabetes efficiently in lymphopenic hosts after conversion into Th1 cells. Eur J Immunol 2009; 39: 216–224.
Lee YK, Turner H, Maynard CL, Oliver JR, Chen DQ, Elson CO et al. Late Developmental Plasticity in the T Helper 17 Lineage. Immunity 2009; 30: 92–107.
Song X, He X, Li X, Qian Y . The roles and functional mechanisms of interleukin-17 family cytokines in mucosal immunity. Cell Mol Immunol 2016; 13: 418–431.
Ferber IA, Brocke S, TaylorEdwards C, Ridgway W, Dinisco C, Steinman L et al. Mice with a disrupted IFN-gamma gene are susceptible to the induction of experimental autoimmune encephalolmyelitis (EAE). J Immunol 1996; 156: 5–7.
Cua DJ, Sherlock J, Chen Y, Murphy CA, Joyce B, Seymour B et al. Interleukin-23 rather than interleukin-12 is the critical cytokine for autoimmune inflammation of the brain. Nature 2003; 421: 744–748.
Yen D, Cheung J, Scheerens H, Poulet F, McClanahan T, Mckenzie B et al. IL-23 is essential for T cell-mediated colitis and promotes inflammation via IL-17 and IL-6. J Clin Invest 2006; 116: 1310–1316.
Ahern PP, Schiering C, Buonocore S, McGeachy MJ, Cua DJ, Maloy KJ et al. Interleukin-23 drives intestinal inflammation through direct activity on T cells. Immunity 2010; 33: 279–288.
Burkett PR, Meyer zu Horste G, Kuchroo VK . Pouring fuel on the fire: Th17 cells, the environment, and autoimmunity. J Clin Invest 2015; 125: 2211–2219.
Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 2009; 139: 485–498.
Gaboriau-Routhiau V, Rakotobe S, Lecuyer E, Mulder I, Lan A, Bridonneau C et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity 2009; 31: 677–689.
Ivanov II, Frutos Rde L, Manel N, Yoshinaga K, Rifkin DB, Sartor RB et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe 2008; 4: 337–349.
Shih VF, Cox J, Kljavin NM, Dengler HS, Reichelt M, Kumar P et al. Homeostatic IL-23 receptor signaling limits Th17 response through IL-22-mediated containment of commensal microbiota. Proc Natl Acad Sci USA 2014; 111: 13942–13947.
Ivanov II, Littman DR . Segmented filamentous bacteria take the stage. Mucosal Immunol 2010; 3: 209–212.
Yang Y, Torchinsky MB, Gobert M, Xiong H, Xu M, Linehan JL et al. Focused specificity of intestinal TH17 cells towards commensal bacterial antigens. Nature 2014; 510: 152–156.
Sano T, Huang W, Hall JA, Yang Y, Chen A, Gavzy SJ et al. An IL-23R/IL-22 Circuit Regulates Epithelial Serum Amyloid A to Promote Local Effector Th17 Responses. Cell 2015; 163: 381–393.
Atarashi K, Nishimura J, Shima T, Umesaki Y, Yamamoto M, Onoue M et al. ATP drives lamina propria T(H)17 cell differentiation. Nature 2008; 455: 808–812.
Killeen ME, Ferris L, Kupetsky EA, Falo L Jr, Mathers AR . Signaling through purinergic receptors for ATP induces human cutaneous innate and adaptive Th17 responses: implications in the pathogenesis of psoriasis. J Immunol 2013; 190: 4324–4336.
Kusu T, Kayama H, Kinoshita M, Jeon SG, Ueda Y, Goto Y et al. Ecto-nucleoside triphosphate diphosphohydrolase 7 controls Th17 cell responses through regulation of luminal ATP in the small intestine. J Immunol 2013; 190: 774–783.
Park J, Kim M, Kang SG, Jannasch AH, Cooper B, Patterson J et al. Short-chain fatty acids induce both effector and regulatory T cells by suppression of histone deacetylases and regulation of the mTOR-S6K pathway. Mucosal Immunol 2015; 8: 80–93.
Sefik E, Geva-Zatorsky N, Oh S, Konnikova L, Zemmour D, McGuire AM et al. MUCOSAL IMMUNOLOGY. Individual intestinal symbionts induce a distinct population of RORgamma(+) regulatory T cells. Science 2015; 349: 993–997.
Ohnmacht C, Park JH, Cording S, Wing JB, Atarashi K, Obata Y et al. MUCOSAL IMMUNOLOGY. The microbiota regulates type 2 immunity through RORgammat(+) T cells. Science 2015; 349: 989–993.
Wing K, Onishi Y, Prieto-Martin P, Yamaguchi T, Miyara M, Fehervari Z et al. CTLA-4 control over Foxp3+ regulatory T cell function. Science 2008; 322: 271–275.
Qureshi OS, Zheng Y, Nakamura K, Attridge K, Manzotti C, Schmidt EM et al. Trans-endocytosis of CD80 and CD86: a molecular basis for the cell-extrinsic function of CTLA-4. Science 2011; 332: 600–603.
Schiering C, Krausgruber T, Chomka A, Frohlich A, Adelmann K, Wohlfert EA et al. The alarmin IL-33 promotes regulatory T-cell function in the intestine. Nature 2014; 513: 564–568.
Pabst O, Herbrand H, Friedrichsen M, Velaga S, Dorsch M, Berhardt G et al. Adaptation of solitary intestinal lymphoid tissue in response to microbiota and chemokine receptor CCR7 signaling. J Immunol 2006; 177: 6824–6832.
Moreau MC, Corthier G . Effect of the gastrointestinal microflora on induction and maintenance of oral tolerance to ovalbumin in C3h-Hej mice. Infect Immun 1988; 56: 2766–2768.
Eberl G, Marmon S, Sunshine MJ, Rennert PD, Choi Y, Littman DR . An essential function for the nuclear receptor RORgamma(t) in the generation of fetal lymphoid tissue inducer cells. Nat Immunol 2004; 5: 64–73.
Sun Z, Unutmaz D, Zou YR, Sunshine MJ, Pierani A, Brenner-Morton S et al. Requirement for RORgamma in thymocyte survival and lymphoid organ development. Science 2000; 288: 2369–2373.
Yokota Y, Mansouri A, Mori S, Sugawara S, Adachi S, Nishikawa S et al. Development of peripheral lymphoid organs and natural killer cells depends on the helix-loop-helix inhibitor Id2. Nature 1999; 397: 702–706.
Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL . An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 2005; 122: 107–118.
Cupedo T, Lund FE, Ngo VN, Randall TD, Jansen W, Greuter MJ et al. Initiation of cellular organization in lymph nodes is regulated by non-B cell-derived signals and is not dependent on CXC chemokine ligand 13. J Immunol 2004; 173: 4889–4896.
Jang MH, Sougawa N, Tanaka T, Hirata T, Hiroi T, Tohya K et al. CCR7 is critically important for migration of dendritic cells in intestinal lamina propria to mesenteric lymph nodes. J Immunol 2006; 176: 803–810.
Wendland M, Willenzon S, Kocks J, Davalos-Misslitz AC, Hammerschmidt SI, Schumann K et al. Lymph node T cell homeostasis relies on steady state homing of dendritic cells. Immunity 2011; 35: 945–957.
Hamada H, Hiroi T, Nishiyama Y, Takahashi H, Masunaga Y, Hachimura S et al. Identification of multiple isolated lymphoid follicles on the antimesenteric wall of the mouse small intestine. J Immunol 2002; 168: 57–64.
Clarke TB . Microbial programming of systemic innate immunity and resistance to infection. PLoS Pathog 2014; 10: e1004506.
Inagaki H, Suzuki K, Nomoto K, Yoshikai Y . Increased susceptibility to primary infection with Listeria monocytogenes in germfree mice may be due to lack of accumulation of L-selectin(+) CD44(+) T cells in sites of inflammation. Infect Immun 1996; 64: 3280–3287.
Robinson CM, Pfeiffer JK . Viruses and the Microbiota. Annu Rev Virol 2014; 1: 55–69.
Moussion C, Girard JP . Dendritic cells control lymphocyte entry to lymph nodes through high endothelial venules. Nature 2011; 479: 542–546.
Zhang Z, Li J, Zheng W, Zhao G, Zhang H, Wang X et al. Peripheral lymphoid volume expansion and maintenance are controlled by gut microbiota via RALDH+ dendritic cells. Immunity 2016; 44: 330–342.
Acknowledgements
YS is supported by the joint Peking-Tsinghua Center for Life Sciences, the National Natural Science Foundation of China General Program (31370878) and by grants from the US NIH (R01AI098995), the Natural Sciences and Engineering Research Council of Canada (RGPIN-355350/396037) and the Canadian Institutes of Health Research (MOP-119295).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Shi, Y., Mu, L. An expanding stage for commensal microbes in host immune regulation. Cell Mol Immunol 14, 339–348 (2017). https://doi.org/10.1038/cmi.2016.64
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cmi.2016.64
Keywords
This article is cited by
-
Reproducible and opposing gut microbiome signatures distinguish autoimmune diseases and cancers: a systematic review and meta-analysis
Microbiome (2022)
-
Commensal viruses maintain intestinal intraepithelial lymphocytes via noncanonical RIG-I signaling
Nature Immunology (2019)
-
T lymphocytes in the intestinal mucosa: defense and tolerance
Cellular & Molecular Immunology (2019)
-
Bridging intestinal immunity and gut microbiota by metabolites
Cellular and Molecular Life Sciences (2019)
-
Dietary therapy may be sufficient for type 1 diabetes treatment
Cellular & Molecular Immunology (2018)