Abstract
Helicobacter pylori (H. pylori) infection is associated with chronic gastritis, peptic ulcer and gastric cancer. Apoptosis induced by microbial infections is implicated in the pathogenesis of H. pylori infection. Here we show that human gastric epithelial cells sensitized to H. pylori confer susceptibility to TRAIL-mediated apoptosis via modulation of death receptor signaling. Human gastric epithelial cells are intrinsically resistant to TRAIL-mediated apoptosis. The induction of TRAIL sensitivity by H. pylori is dependent on the activation of caspase-8 and its downstream pathway. H. pylori induces caspase-8 activation via enhanced assembly of the TRAIL death-inducing signaling complex (DISC) through downregulation of cellular FLICE-inhibitory protein (FLIP). Overexpression of FLIP abolished the H. pylori-induced TRAIL sensitivity in human gastric epithelial cells. Our study thus demonstrates that H. pylori induces sensitivity to TRAIL apoptosis by regulation of FLIP and assembly of DISC, which initiates caspase activation, resulting in the breakdown of resistance to apoptosis, and provides insight into the pathogenesis of gastric damage in Helicobacter infection. Modulation of host apoptosis signaling by bacterial interaction adds a new dimension to the pathogenesis of Helicobacter.
Similar content being viewed by others
Main
Helicobacter pylori, a common human pathogen that infects about 50% of the world’s population, is associated with duodenal ulcer and peptic ulcer diseases. Recent studies have shown that there is increased apoptosis of gastric epithelial cells during H. pylori infection.1, 2, 3 Enhanced gastric epithelial cell apoptosis observed during infection with H. pylori is believed to be significant in the etiology of gastritis, peptic ulcers, and neoplasia. Recent studies have suggested that T cells are selectively increased during infection.4, 5, 6, 7 Cytokines like gamma interferon (IFN-γ) and tumor necrosis factor alpha (TNF-α) can increase the release of proinflammatory cytokines and augment apoptosis induced by H. pylori.8 H. pylori infection could also induce damage to gastric mucosa by increasing the expression of Fas in gastric epithelial cells, leading to gastric epithelial cell apoptosis through Fas/FasL interaction with infiltrating T cells.9, 10 These findings suggest a role for immune-mediated apoptosis of gastric epithelial cells during H. pylori infection.
TRAIL (tumor necrosis factor-related apoptosis-inducing ligand; also called Apo2L), a novel TNF superfamily member with a strong homology to FasL, is capable of inducing apoptosis in a variety of transformed cell lines in vitro11, 12 but usually not in normal primary cells. It was recently shown that T cells kill target cells via TRAIL/TRAIL receptor interaction,13, 14, 15 suggesting that TRAIL might serve as a cytotoxic effector molecule in activated T cells in vivo. Recent studies indicate that TRAIL-induced apoptosis occurs through a caspase signaling cascade and that resistance to TRAIL is controlled by intracellular regulators of apoptosis. The molecular events leading to apoptosis induction via TRAIL have recently been analyzed. Crosslinking of the TRAIL receptors leads to the formation of a death-inducing signaling complex (DISC).16 The death adaptor protein FADD (MORT1) and the proteolytic enzymes caspase-8 and -10 are recruited to the TRAIL DISC.17, 18, 19, 20, 21 Procaspase-8 is proteolytically cleaved and activated at the DISC. Activated caspase-8 then initiates the apoptosis executing caspase cascade. Previous studies demonstrated that TRAIL-induced apoptosis could be enhanced by Helicobacter and hepatitis C virus core protein.22, 23 However, the mechanisms leading to induce TRAIL sensitivity by microbes are not clear. Upregulation of the apoptosis-inducing TRAIL receptors after treatment with chemotherapeutic drugs and radiation has been implicated in sensitizing human leukemic and glioma cells.24, 25, 26 It has been shown that chemotherapy-resistant tumor cells can be sensitized for TRAIL-induced apoptosis at the DISC level.27 Here we report that human gastric epithelial cells sensitized to H. pylori confer susceptibility to TRAIL-mediated apoptosis. H. pylori induces TRAIL apoptosis signaling by downregulation of FLICE-inhibitory protein (FLIP), which enhances the assembly of TRAIL DISC, induces caspase-8 activation, and conveys the death signal to mitochondria, resulting in a breakdown of apoptosis resistance.
Results
H. pylori enhances sensitivity to TRAIL-mediated apoptosis in human gastric epithelial cell lines via activation of caspase-8 and its downstream pathway
TRAIL has been shown to induce apoptosis in a number of different tumor cell types but not usually in normal primary cells. To examine a role for TRAIL-induced apoptosis in gastric epithelial cells, recombinant TRAIL proteins were used to induce apoptosis in human gastric epithelial cell lines (AGS). The results revealed that AGS cells were resistant to TRAIL-mediated apoptosis. We further studied TRAIL-induced apoptosis in gastric epithelial cells after interaction with H. pylori. In the absence of TRAIL, H. pylori induced only mild apoptosis in AGS cells; however, apoptosis was markedly induced after adding TRAIL, and induction of TRAIL-mediated apoptosis by H. pylori was specifically blocked by adding soluble TRAIL receptor, death receptor 4 (DR4)-Fc, indicating that cell death resulted from the interaction between TRAIL and the TRAIL receptor on the cell surface (Figure 1a).
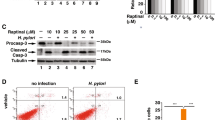
H. pylori enhances sensitivity to TRAIL-mediated apoptosis in human gastric epithelial cell lines via activation of caspase-8 and its downstream pathway. (a) Human gastric epithelial cell line AGS were co-cultured with H. pylori for 12 h and incubated with recombinant TRAIL proteins in the presence or absence of soluble TRAIL receptor, DR4-Fc protein, for 12 h. Cell apoptosis was measured by cell death enzyme-linked immunosorbent assay (ELISA). Values represent means±S.D. of three independent experiments (*P<0.05). (b) AGS cells were co-cultured with H. pylori for 12 h and incubated with recombinant TRAIL proteins for the indicated times. Cell lysates were subjected to western blotting analysis for detection of caspase activation. The native forms of caspase-8 and caspase-3 are 55/54 and 32 kDa, respectively. The cleaved forms of caspase-8 are 43/41 and 18 kDa, respectively. The cleaved forms of caspase-3 are 19 and 17 kDa, respectively. The expression of actin serves as a loading control. CASP, caspase. (c) AGS cells were co-cultured with H. pylori for 12 h in the presence of pan-caspase inhibitor, Z-VAD-fmk (Bachem, Bubendorf, Sweden), specific caspase-8 inhibitor, Z-IETD-fmk, or specific caspase-3 inhibitor, Z-DEVD-fmk (Calbiochem, San Diego, CA, USA) and then were treated with recombinant TRAIL proteins for 4 h. Cell apoptosis was measured by cell death ELISA. Values represent means±S.D. of three independent experiments (*P<0.05). (d) AGS cells were transfected with scramble or caspase-8 siRNA, followed by incubating with H. pylori for 12 h and subsequently treated with TRAIL for 4 h. Cell apoptosis was measured by cell death ELISA. Values represent means±S.D. of three independent experiments (*P<0.05)
To further delineate the intracellular signal transduction pathway modulated by H. pylori that results in induction of TRAIL-sensitivity, we investigated the activation of caspase pathways following TRAIL engagement and subsequent to H. pylori interaction. In the absence of H. pylori, TRAIL engagement only induced a slight activation of caspase-8 but not caspase-3 in AGS cells. In contrast, during TRAIL engagement, both caspase-8 and caspase-3 were fully activated in AGS cells, resulting in cell apoptosis after exposure to H. pylori (Figure 1b). Furthermore, the ability to induce TRAIL sensitivity in AGS cells by H. pylori was significantly suppressed by the caspase-8 inhibitor, Z-IETD-fmk; pan-caspase inhibitor, Z-VAD-fmk; or the caspase-3 inhibitor, DEVD-fmk. (Figure 1c) In addition, the H. pylori-induced TRAIL sensitivity in AGS cells was significantly suppressed by knocking down caspase-8 (Figure 1d).
To exclude the possibility that the H. pylori-induced sensitivity to TRAIL in gastric epithelial cell lines is due to upregulation of TRAIL DRs, we further examined the expression of TRAIL receptors before and after interaction with H. pylori. The results demonstrated that expression of TRAIL receptors on the gastric cell surface did not appear significantly different before and after interaction with H. pylori (Figure 2). Taken together, the results indicate that H. pylori enhances TRAIL-mediated apoptosis in gastric epithelial cell lines by modulating intracellular death signal transduction by activation of a caspase-8 downstream cascade. In so doing, the pathogen alters the intracellular regulation of resistance to DR-induced apoptosis through a pathway involving the sequential induction of apical caspase-8 activity, caspase cascade, and effector caspase-3 activity.
H. pylori-induced TRAIL sensitivity in gastric epithelial cells is not due to upregulation of DRs. AGS Cells were incubated with or without H. pylori for 12 h and then the expression of TRAIL receptors, DR4, DR5, DcR1, and DcR2, on cell surfaces were analyzed by flow cytometry with mAbs against DR4, DR5, DcR1, and DcR2 (eBioscience) or isotype antibody (shaded histogram). The human granulocytes served as positive controls for DcR1 and DcR2
H. pylori induces the activation of the mitochondrial signaling pathway after TRAIL engagement
For detecting caspase processing events distal to caspase-8 activation, we investigated the activation of mitochondria after exposure to H. pylori in AGS cells. To examine the changes of mitochondrial membrane potential (Δψm) after TRAIL engagement, the Δψm was detected by the uptake of 5,5′,6,6′-tetrachloro-1,1′,3,3′-tetrathylbenzimidazolyl carbocyanine iodide (JC-1) dye.28, 29 The results (Figure 3) demonstrate that TRAIL engagement induced a breakdown in the transmembrane potential of mitochondria after interaction with H. pylori but not in the absence of H. pylori, consistent with the difference in caspase-3 activation. To further confirm the induction of mitochondrial signaling pathway by TRAIL, we also analyzed cytochrome c released from mitochondria into cytoplasm after TRAIL engagement. The results (Figure 3) demonstrate that TRAIL engagement induced the release of cytochrome c from mitochondria to cytosol and activated the mitochondria downstream caspase cascade, caspase-9, in AGS cells in the presence of H. pylori. In addition, the induction of TRAIL sensitivity in AGS cells by H. pylori was significantly inhibited in the presence of the caspase-9 inhibitor Z-LEHD-fmk (Figure 3d), indicating that activation of the mitochondria downstream pathway was required in TRAIL-mediated apoptosis in AGS cells induced by H. pylori.
H. pylori induces activation of the mitochondrial pathway after TRAIL engagement. (a and b) Changes in mitochondrial transmembrane potential were assessed as JC-1 green (uncoupled mitochondria) or red (intact mitochondria) fluorescence. Green fluorescence of JC-1 dye was assessed as a parameter for mitochondrial breakdown. (a) AGS cells were labeled with the mitochondrial transmembrane potential-sensitive dye JC-1 and treated with recombinant TRAIL in the presence or absence of H. pylori. The JC-1 green (uncoupled mitochondria) or red (intact mitochondria) fluorescence was visualized under a confocal microscope. Reduced red fluorescence with increased green fluorescence was observed in AGS cells treated with TRAIL in the presence of H. pylori. There were 100 cells analyzed per field by confocal microscopy, and this is a representative sample. (b) AGS cells were labeled with JC-1 and treated with recombinant TRAIL in the presence or absence of H. pylori. Analysis was performed by a FACS scan. Increased green fluorescence was detectable in AGS cells treated with recombinant TRAIL after exposure to H. pylori (thick line) and compared with AGS cells treated with TRAIL but without exposure to H. pylori (thin line). Shaded histogram: AGS alone. (c) H. pylori induces the release of cytochrome c from mitochondria after TRAIL engagement. AGS cells (106 cells) were incubated with 1 g/ml recombinant TRAIL proteins for various times as indicated in the absence or presence of H. pylori, and the cytoplasmic fraction of the cell lysate was isolated. Cytoplasmic proteins were separated by SDS-PAGE and then transferred to a nitrocellulose membrane. Cytochrome c released from mitochondria into the cytoplasm was detected with specific anti-cytochrome c and anti-caspase-9 mAbs (Cell Signaling Technology). The native form of caspase-9 is 47 kDa, and cleaved forms are 37 kDa and 35 kDa. Membranes were also blotted with anti-β-actin mAb as a loading control. The results were representative of at least three independent experiments. (d) AGS Cells were co-cultured with H. pylori and treated with caspase-9 inhibitor, Z-LEHD-fmk, followed by treatment with TRAIL proteins. Cell apoptosis was measured by cell death enzyme-linked immunosorbent assay (ELISA). Values represent means±S.D. of three independent experiments (*P<0.05). For specificity control of caspase-9 inhibition by Z-LEHD-fmk, the H9 cells, a type I Fas signaling cells, were treated with FasL (Peprotech, Rocky Hill, NJ, USA), a death stimulus that does not require caspase-9, and Etoposide, a death stimulus that requires caspase-9. Cell apoptosis was measured by cell death ELISA. Values represent means±S.D. of three independent experiments (*P<0.05)
To investigate the role of the mitochondria apoptosis signaling pathway in H. pylori-induced TRAIL sensitivity, we further transfected Bcl-2 into AGS cells to establish stable transfectants. The results (Figure 4a) demonstrate that the overexpression of Bcl-2 in AGS cells inhibited the induction of TRAIL sensitivity in AGS cells by H. pylori. Taken together, our results indicate that H. pylori induces a sensitivity to TRAIL-mediated apoptosis in gastric epithelial cell lines through a pathway involving the activation of caspase-8 downstream signaling pathway to convey the death signal to mitochondria, leading to activation of the mitochondrial apoptosis signaling pathway and eliminating resistance to apoptosis.
H. pylori-induced TRAIL sensitivity is dependent on Bid processing. (a) AGS cells were transfected with Bcl-2, followed by incubation with H. pylori and subsequently treated with TRAIL. Cell apoptosis was measured by cell death enzyme-linked immunosorbent assay (ELISA). Values represent means±S.D. of three independent experiments (*P<0.05). (b) AGS cells were incubated with or without H. pylori followed by treatment with TRAIL at the indicated times. The expression of full-length and truncated Bid (22 and 15 kDa, respectively) was detected by western blotting. Membranes were also blotted with anti-β-actin mAb as a loading control. (c) Cells were transfected with scramble or Bid siRNA, followed by incubation with H. pylori and subsequently treated with TRAIL. Cell apoptosis was measured by cell death ELISA. Values represent means±S.D. of three independent experiments (*P<0.05)
H. pylori induces sensitivity to TRAIL-mediated apoptosis and activates the mitochondrial signaling pathway via Bid cleavage after TRAIL engagement
The finding that TRAIL engagement induces cytochrome c release and caspase-9 activation in AGS cells in the presence of H. pylori indicates that apoptosis signals are conveyed from the TRAIL DR to mitochondria in AGS cells after interaction with H. pylori. For detecting the caspase processing events distal to caspase-8, which connects caspase-8 with mitochondria, we further investigated the processing of Bid after TRAIL engagement. The results (Figure 4b) demonstrate that H. pylori enhanced TRAIL-induced Bid cleavage in AGS cells, which is consistent with activation of the mitochondrial signaling pathway and caspase-3 after TRAIL engagement. In addition, the H. pylori-induced TRAIL sensitivity in AGS cells was significantly suppressed by knocking down Bid (Figure 4c). Taken together, our results indicate that the induction of TRAIL sensitivity by H. pylori is dependent on the sequential activation of the caspase-8 downstream pathway, enhancing the cleavage of Bid, activating the mitochondrial signaling pathway, and breaking resistance to apoptosis.
H. pylori enhances the assembly of the TRAIL DISC through reduction of FLIP recruitment
The expression of DR4 and DR5 DRs was not upregulated by H. pylori, and results indicate that H. pylori enhances TRAIL-mediated apoptosis in gastric epithelial cell lines by modulating intracellular death signal transduction. We then investigated the possible involvement of FLIP in regulating the assembly of TRAIL DISC in the presence or absence of H. pylori. Cells were co-cultured with H. pylori and then stimulated with FLAG-TRAIL. After TRAIL engagement, the cell lysate was co-immunoprecipitated with an anti-FLAG monoclonal antibody (mAb), and the isolated receptor complexes were analyzed for TRAIL DISC via western blotting. The results (Figure 5) demonstrate that H. pylori enhanced the activation of caspase-8 during TRAIL engagement in a dose-dependent manner in human gastric epithelial cells (Figure 5a). When we investigated the assembly of the TRAIL DISC after TRAIL engagement in the presence or absence of H. pylori via co-immunoprecipitation experiments, the results (Figure 5b) demonstrate that, in the absence of H. pylori, TRAIL induced the assembly of DR5 (TRAIL-R2), FADD (MORT1), FLIP, and a small amount of caspase-8 in the DISC. In contrast, H. pylori significantly enhanced the recruitment of caspase-8 into the TRAIL DISC through reduction of the FLIP assembly and therefore augmented the activation of caspase-8. Our results indicate that H. pylori enhances the assembly of the TRAIL DISC and activation of caspase-8 via inhibition of FLIP recruitment into the TRAIL DISC.
H. pylori downregulates FLIPS expression and enhances the TRAIL-induced DISC assembly. (a) AGS cells were incubated with increasing amounts of H. pylori (MOI 25, 50, 100, 200) for 12 h and subsequently treated with TRAIL. Cell lysates were analyzed by western blotting to detect the activation level of caspase-8. Actin expression was used as a protein loading control. (b) Cells were co-cultured with H. pylori for 12 h, stimulated with 1 μg/ml FLAG-TRAIL combined with 2 μg/ml anti-FLAG antibody or normal mouse IgG for 1 h, and cell lysates were precipitated with protein A agarose. Isolated receptor complexes (IP) were assessed for caspase-8, FLIPS, FADD, and DR5 by western blotting
Sensitization of TRAIL-induced apoptosis by H. pylori is mediated by downregulation of c-FLIPS
Our results indicate that H. pylori enhances the assembly of the TRAIL DISC by inhibiting the recruitment of FLIP into the DISC. To further confirm the role of FLIP in H. pylori-induced TRAIL sensitivity in human gastric epithelial cells, AGS cells were transfected with FLIPS, co-cultured with H. pylori, and then subjected to treatment with TRAIL. The results (Figure 6) demonstrate that TRIAL sensitivity induced by H. pylori is completely abolished by FLIPS overexpression. In addition, the amount of the FLIPS protein was decreased when cells interacted with H. pylori. In contrast, when cells were pretreated with a proteosome inhibitor MG132 for 30 min before the incubation of H. pylori, the decrease in c-FLIPS protein was inhibited (Figure 6b), indicating that H. pylori induced the degradation of FLIPS, leading to a decrease in the recruitment of FLIP into the DISC.
H. pylori-induced sensitization to TRAIL-induced apoptosis is via c-FLIPS downregulation. (a) AGS cells were transfected with FLIPS and pCMV mock control, followed by incubation with H. pylori and subsequently treated with TRAIL. Cell apoptosis was measured by cell death enzyme-linked immunosorbent assay. Values represent means±S.D. of three independent experiments (*P<0.05). (b) Cells were pretreated with 2 μM MG132, a proteosome inhibitor, for 30 min before incubation with H. pylori (MOI 50,100, 200). After co-culture for 12 h, c-FLIPS was determined by western blotting. The expression of actin serves as a loading control
To further confirm the role of FLIP in regulating H. pylori-induced TRAIL sensitivity in human gastric epithelial cells, AGS cells were transfected with c-FLIPS siRNA to knock down the expression of FLIPS and co-cultured with H. pylori followed by treatment with TRAIL. The results (Figure 7) demonstrate that the knockdown of endogenous c-FLIPS markedly enhances TRAIL-induced apoptosis. In addition, the knockdown of c-FLIPS induced the cleavage of caspase-8, Bid, caspase-9, and caspase-3 after TRAIL engagement. Taken together, our results indicated that H. pylori enhances TRAIL sensitivity in human gastric epithelial cells via a decrease in FLIP recruitment into the DISC, leading to the activation of caspase-8 and its downstream pathway, thus breaking TRAIL resistance.
Knockdown of endogenous c-FLIPS enhances TRAIL-mediated apoptosis in human gastric epithelial cells. (a) AGS cells were transfected with scramble or c-FLIPS siRNA, followed by treatment with TRAIL. Cell apoptosis was measured by cell death enzyme-linked immunosorbent assay. Values represent means±S.D. of three independent experiments (*P<0.05). (b) AGS cells were transfected with scramble or c-FLIPS siRNA, followed by treatment with TRAIL. The cell lysate was subjected to western blotting analysis for detection of caspase activation. The expression of actin serves as a loading control
Discussion
Our studies indicate that H. pylori induces TRAIL apoptosis signaling by downregulating FLIP, which enhances the assembly of DISC, induces maximal caspase-8 activation and conveys the death signal to mitochondria, leading to a breakdown in apoptosis resistance. Collectively, our findings uncover a novel mechanism of modulation of TRAIL resistance mediated by H. pylori through regulation of the DISC and provides strong evidence that H. pylori modulates the transduction of the host cell death signal via downregulation of FLIP.
We demonstrated in previous reports that human gastric epithelial cells sensitized to H. pylori confer susceptibility to TRAIL-mediated apoptosis, suggesting a role for immune-mediated apoptosis of gastric epithelial cells by infiltrating T cells during Helicobacter infection.6, 22 The induction of TRAIL sensitivity by H. pylori is independent of the expression of H. pylori virulent factors Vac A and Cag A and is dependent on viable bacteria and direct contact with cells.22 The role of TRAIL DR DR5 (TRAIL-R2) in the regulation of TRAIL-induced apoptosis has been evaluated recently in several tumor cell lines.30, 31, 32, 33, 34 In our study, the expression of TRAIL receptors did not change after H. pylori infection, indicating that the H. pylori-induced enhanced sensitivity to TRAIL-mediated apoptosis in gastric epithelial cells is not due to upregulation of TRAIL DRs, DR5 in particular (Figure 2). Thus, instead of the induction of DR upregulation, H. pylori induces TRAIL sensitivity in gastric epithelial cells via modulation of DR signal transduction pathways.
H. pylori induces sensitivity to TRAIL-mediated apoptosis in gastric epithelial cells through a pathway involving the sequential induction of apical caspase-8 and effector caspase-3 activity via activation of a mitochondrial pathway. Our results suggest that the type II pathway is mainly involved in TRAIL-induced apoptosis in human gastric epithelial cells where mitochondria have an important role in amplifying apoptotic signals. Our results also indicate that H. pylori induces the mitochondrial apoptosis pathway via FLIP downregulation, which enhances DISC assembly after TRAIL engagement and further induces the activation of caspase-8 and cleavage of Bid. c-FLIP proteins have an essential role in the regulation of DR-induced apoptosis. Among the three isoforms: c-FLIP long (c-FLIPL, 55 kDa), c-FLIP short (c-FLIPS, 27 kDa), and c-FLIP Raji (c-FLIPR, 25 kDa),35, 36, 37 c-FLIPS is a well-described inhibitor of DR-mediated apoptosis.35, 36, 37, 38 It has been shown that c-FLIPS blocks Fas35, 36, 37, 38 as well as TRAIL35 induced apoptosis via binding to the DISC and inhibiting procaspase-8 processing and activation at the DISC.35, 36, 37, 38 However, the effect of the c-FLIPL isoform on DR-mediated apoptosis is still controversial. Our results demonstrated that H. pylori modulated the amounts of c-FLIPS; and overexpression of c-FLIPS abolished the effects of H. pylori-induced TRAIL sensitivity in AGS cells. Therefore, our study indicates that short c-FLIPS isoform modulates TRAIL apoptosis signaling in human gastric epithelial cells.
During H. pylori infection, the degree of apoptosis induced in the stomach is affected by the inflammatory response. H. pylori infection induces a T-cell response and a number of inflammatory mediators, including cytokines and chemokines.4, 5, 6, 7, 39, 40 These infiltrating T cells can then kill gastric target cells via TRAIL/TRAIL receptor interaction. In addition to its role in inducing apoptosis by binding to DRs, TRAIL itself can stimulate T cell after T-cell receptor (TCR) engagement and augment IFN-γ secretion.41, 42, 43 Our results, combined with recent evidence, therefore support the functional role of TRAIL in H. pylori-induced apoptosis. However, the degree of apoptosis during gastric inflammation is linked to the presence of H. pylori and the associated inflammatory response. Therefore, the degree of mucosal damage not only depends on the virulence of H. pylori but is also determined by the inflammatory response that is induced by H. pylori within the gastric epithelium.
Recently, several bacterial pathogens have been found to trigger apoptosis in host cells in vitro or in vivo, and several types of mechanisms have been elucidated.44 Recent reports have shown that H. pylori can secrete CagA into gastric epithelial cell by Type IV secretion, inducing intracellular protein phosphorylation in host cells,45, 46, 47, 48 suggesting that H. pylori can modulate intracellular signaling via factors secreted into host cells. However, in our previous study we demonstrated that induction of TRAIL sensitivity by H. pylori is independent of H. pylori virulent factors Vac A and Cag A.22 Therefore, the bacterial factors leading to induction of TRAIL sensitivity by H. pylori is still not clear, and it awaits further investigation. Taken together, our results indicate that H. pylori modulates the sensitivity to TRAIL-mediated apoptosis in gastric epithelial cells by the induction of FLIP downregulation, which enhances DISC assembly, further induces caspase-8 activation, and conveys the death signal to mitochondria, resulting in a breakdown of apoptosis resistance and causing gastric mucosal damage during inflammation. Thus, modulation of host cell death signal transduction by bacterial interaction adds a new dimension to the immune pathogenesis in chronic Helicobacter infection.
Materials and Methods
Bacterial strains, cell lines, and media
H. pylori strains ATCC 43504 were obtained from American Type Culture Collections (ATCC, Manassas, VA, USA). Before each experiment, H. pylori was passaged on 5% sheep blood agar plates by incubation in an atmosphere consisting of 5% O2, 15% CO2, and 80% N2 for 2–4 days at 37 °C. Bacteria were cultured in brucella broth (Difco Labs, Detroit, MI, USA) supplemented with 5% FBS, vancomycin and amphotericin B under the same condition for 30 h at 37 °C with agitation (80–100 r.p.m.). Cells were then pelleted and resuspended in phosphate-buffered saline at a concentration of 108 CFU/ml. The human gastric adenocarcinoma cell lines AGS were obtained from ATCC and maintained in Dulbecco modified essential medium (DMEM), supplemented with 10% FBS.
Expression and purification of recombinant TRAIL protein and soluble TRAIL receptor DR4-Fc
The recombinant TRAIL proteins were expressed in the Escherichia coli expression system and purified with a Ni column.41 In brief, the coding portion of the extracellular domain of TRAIL (amino acids 123–314) was PCR-amplified, subcloned into pRSET B vector (Invitrogen, Groningen, Netherlands), and expressed in the E. coli expression system. The purification of recombinant His-TRAIL fusion protein was performed by metal chelate column chromatography using Ni-NTA resin according to the manufacturer’s recommendations (Qiagen, Hilden, Germany). His-TRAIL was quantified by the Bradford method and protein assay reagent (BioRad, Richmond, CA, USA). To generate soluble recombinant DR4-Fc fusion molecules, the coding sequence for the extracellular domain of human DR4 was isolated by RT-PCR using the forward primer, 5′-CGGATTTCATGGCGCCACCACCA-3′ and the reverse primer, 5′-GAAGATCTATTATGTCCATTGCC-3′. The amplified product was ligated in-frame into BamHI-cut pUC19-IgG1-Fc vector containing the human IgG1 Fc coding sequence. The fusion gene was then subcloned into pBacPAK9 vector (Clontech, Palo Alto, CA, USA). DR4-Fc fusion protein was recovered from the filtered supernatants of the recombinant virus-infected Sf21 cells using protein G-Sepharose beads (Pharmacia, Piscataway, NJ, USA). The bound DR4-Fc protein was eluted with glycine buffer (pH 3) and dialyzed into PBS.
Apoptosis assay
A sensitive ELISA assay, which detects cytoplasmic histone-associated DNA fragments, was performed according to the manufacturer’s protocol (Cell Death Detection ELISAPLUS; Roche Mannheim Biochemicals, Mannheim, Germany). This was cultured overnight in a 96-well plate at 104 gastric epithelial cells/well, treated with TRAIL recombinant protein for 4 h, and harvested by centrifugation at 200 × g. Cells were lysed by incubation with lysis buffer for 30 min, followed by centrifugation at 200 × g for 10 min at room temperature. The supernatant was collected and incubated with immunoreagent that was prepared 2 h beforehand. After washing gently, the supernatant was pipetted into each well along with the substrate solution and kept in the dark until it developed sufficient color for photometric analysis. The reaction was determined in a spectrophotometer at 405 nm. Caspase inhibitors were used in some experiments. Caspase-8, caspase-3, or caspase-9 inhibitors were applied at a concentration of 10 μM to the medium 30 min before treatment.
Flow cytometry
AGS cells were resuspended in 90 μl FACS staining buffer (1% FCS in PBS), incubated with various fluorochrome-conjugated antigen-specific antibodies (anti-DR4, -DR5, -DcR1 (decoy receptor 1) and -DcR2) (eBioscience, San Diego, CA, USA) at 4 °C for 30 min, and followed by washing twice with 1 ml of FACS staining buffer. Cell fluorescence intensity was detected by a FACScan and analyzed by the CellQuest software.
Detection of Δψm
To detect the changes in ΔΨm, AGS cells were seeded at 106 cells/ml into a 6-well plate and grown in DMEM supplemented with 10% fetal calf serum in a 95% air–5% CO2 atmosphere at 37 °C in a humidified incubator overnight. Cells were co-cultured with HP strain 51932 at a concentration of 5 × 108 CFU/ml for 12 h and treated with 1.5 μg/ml TRAIL recombinant protein for another 12 h. Mitochondrial injury and changes in the Δψm were assessed by staining with JC-1 (Molecular Probes, Eugene, OR, USA). This dye, existing as a monomer in solution, emits a green fluorescence and can assume a dimeric configuration that emits red fluorescence in a reaction that is driven by the mitochondrial transmembrane potential.28 Thus, red fluorescence indicates intact mitochondria, whereas green fluorescence shows monomeric JC-1 that remained unprocessed due to breakdown of the mitochondrial transmembrane potential. After trypsinization by TEG buffer (0.125% trypsin, 0.05% EDTA, 0.05% glucose in PBS), the cells were resuspended in DMEM and incubated with JC-1 10 μg/ml for 15 min at 37 °C. Cells were washed with PBS twice and resuspended in PBS. Analysis was performed by a FACS scan, and mitochondrial function was assessed as JC-1 green (uncoupled mitochondria) or red (intact mitochondria) fluorescence.29 For confocal laser scanning microscopy analysis of mitochondrial function, the JC-1-treated cells were excited at 488 nm, and emissions were simultaneously recorded at 527 and 590 nm into an independent detector.
Preparation of cytosols, analysis of cytochrome c release, and Bid cleavage
After incubation with TRAIL, cells were washed twice with ice-cold PBS. They were suspended in 100 μl of extraction buffer (50 mM PIPES-KOH, pH 7.4, 220 mM mannitol, 68 mM sucrose, 50 mM KCl, 5 mM EGTA, 2 mM MgCl2, 1 mM dithiothreitol, and protease inhibitors) and allowed to swell on ice for 30 min. Cells were homogenized by passing the suspension through a 25-gauge needle (10 strokes). Homogenates were centrifuged at 14 000 × g for 15 min at 4 °C, and the supernatants were harvested and stored at −80 °C. The protein content of the cytosols was determined by the Bio-Rad protein assay (Bio-Rad Laboratories, Hercules, CA, USA). For the analysis of cytochrome c release, 10 μg of cytosolic protein was loaded per lane. Proteins were separated on 12% SDS-PAGE and transferred to nitrocellulose sheets, which were blocked for 1 h in PBS and 0.05% Tween 20 with 5% dry milk. Blots were probed in PBS and 0.05% Tween 20, with anti-cytochrome c mAb (eBioscience) or anti-Bid polyclonal Ab (Cell Signaling, Beverly, MA, USA) and secondary antibodies. The immunoblots were checked with anti-β actin mAb to ensure equal loading.
Immunoblot analysis
Cells (5 × 105/ml) were washed twice with ice-cold PBS, scraped from their 6-cm culture dishes into 1 ml ice-cold PBS, and centrifuged at 1000 × g for 5 min. The resultant pellets were incubated with RIPA lysis buffer (150 mM NaCl, 50 mM Tris-HCl (pH 8.0), 1% NP40, 0.1% SDS, 0.5% sodium deoxycholateor, 10 mg/ml phenylmethylsulfonyl fluoride, and 10 mg/ml aprotinin) for 30 min on ice. The cell lysates were centrifuged, and the supernatant was stored at −80 °C until used. The protein concentrations of the extracts were measured with protein assay reagent. The extracted cell lysates (50 μg) were subjected to SDS-PAGE and transferred onto PVDF. The membrane was blocked in 5% nonfat skim milk or 5% BSA in TBST (100 mM Tris-HCl at pH 7.4, 9% NaCl, 2% Tween-20) at room temperature for 30 min and incubated with mouse mAb against caspase-3 (Imgenex, San Diego, CA, USA), caspase-8 (Cell Signaling Technology, Danvers, CO, USA), FLIP (ENZO Life Sciences, Farmingdale, NY, USA), and actin (Chemicon International, Temecula, CA, USA) or rabbit polyclonal antibody against caspase-9, cytochrome c, Bid (Cell Signaling Technology), FADD (ENZO Life Sciences), and DR5 (Chemicon International). Bound primary antibodies were detected with an anti-mouse IgG or anti-rabbit IgG conjugated with HRP (Cell Signaling Technology), and the bands were visualized using the enhanced chemiluminescence method (Perkin-Elmer, Waltham, MA, USA). The expression of actin was used to verify equal loading in all studies.
DISC immunoprecipitation
For co-immunoprecipitation of TRAIL DISC, 5 × 106 AGS cells were used per sample. Cells were seeded onto 10-cm dishes, incubated overnight with H. pylori cells for 12 h, and harvested at 1300 r.p.m. for 5 min. Cells were resuspended with 500 μl RPMI medium, treated with 0.5 μg Flag-TRAIL (ENZO Life Sciences) combined with 1 μg anti-Flag mAb, M2 (Sigma-Aldrich, St. Louis, MO, USA) or normal mouse IgG (Santa Cruz Biotechnology, Santa Cruz, CA, USA), and the mixture was incubated on an end-over-end rotator at room temperature for 1 h. PBS (5 ml) was added to the mixture with 10 × volume of ice, and the cells were collected by centrifugation at 1300 r.p.m. at 4 °C. The cells were lysed with 1000 μl cold lysis buffer with 10 μM Z-VAD-fmk for 30 min at 4 °C. The resulting protein complexes were precipitated from the lysates by adding protein A agarose and then incubated for 1 h at 4 °C. Each sample was washed three times with lysis buffer at 4 °C. Isolated receptor complexes (IP) were assessed for caspase-8, FLIPS, FADD, and DR5 by western blotting.
Abbreviations
- DR:
-
death receptor
- DcR:
-
decoy receptor
- DISC:
-
death-inducing signaling complex
- FLIP:
-
FLICE-inhibitory protein
- IFN-γ:
-
gamma interferon
- mAb:
-
monoclonal antibody
- TRAIL:
-
tumor necrosis factor-related apoptosis-inducing ligand
References
Jones NL, Shannon PT, Cutz E, Yeger H, Sherman PM . Increase in proliferation and apoptosis of gastric epithelial cells early in the natural history of Helicobacter pylori infection. Am J Pathol 1997; 151: 1685–1703.
Moss SF, Calam J, Agarwal B, Wang S, Holt PG . Induction of gastric epithelial apoptosis by Helicobacter pylori. Gut 1996; 38: 498–501.
Rudi J, Kuck D, Strand S, Von Herbay A, Mariani SM, Krammer PH et al. Involvement of the CD95 (APO-1/Fas) receptor and ligand system in Helicobacter pylori-induced gastric epithelial apoptosis. J Clin Invest 1998; 102: 1506–1514.
Bamford KB, Fan XJ, Crowe SE, Leary JF, Gourley WK, Luthra GK et al. Lymphocytes in the human gastric mucosa during Helicobacter pylori have a T helper cell 1 phenotype. Gastroenterology 1998; 114: 482–492.
Elios MM, Manghetti M, De Carli M, Costa F, Baldari CT, Burroni D et al. T helper 1 effector cells specific for Helicobacter pylori in gastric antrum of patients with peptic ulcer disease. J Immunol 1997; 158: 962–967.
Wu YY, Tsai HF, Lin WC, Hsu PI, Shun CT, Wu MS et al. Upregulation of CCL20 and recruitment of CCR6+ gastric infiltrating lymphocytes in Helicobacter pylori gastritis. Infect Immun 2007; 75: 4357–4363.
Tsai HF, Hsu PN . Interplay between Helicobacter pylori and immune cells in immune pathogenesis of gastric inflammation and mucosal pathology. Cell Mol Immunol 2010; 7: 255–259.
Wagner S, Beil W, Westermann J, Logan RPH, Bock CT, Trautwein C et al. Regulation of epithelial cell growth by Helicobacter pylori: evidence for a major role of apoptosis. Gastroenterology 1997; 113: 1836–1847.
Jones N, Day A, Jennings HA, Sherman PM . Helicobacter pylori induces gastric epithelial cell apoptosis in association with increased Fas receptor expression. Infect Immun 1999; 67: 4237–4242.
Wang J, Fan X, Lindholm C, Bennett M, O’Connoll J, Shanahan F et al. Helicobacter pylori modulates lymphoepithelial cell interactions leading to epithelial cell damage through Fas/Fas ligand interactions. Infect Immun 2000; 68: 4303–4311.
Wiley SR, Schooley K, Smolak PJ, Din WS, Huang CP, Nicholl JK et al. Identification and characterization of a new member of the TNF family that induces apoptosis. Immunity 1995; 3: 673–682.
Griffith TS, Chin WA, Jackson GC, Lynch DH, Kubin MZ . Intracellular regulation of TRAIL-induced apoptosis in human melanoma cells. J Immunol 1998; 161: 2833–2840.
Thomas WD, Hersey P . TNF-related apoptosis-inducing ligand (TRAIL) induces apoptosis in Fas ligand-resistant melanoma cells and mediates CD4 T cell killing of target cells. J Immunol 1998; 161: 2195–2200.
Kayagaki N, Yamaguchi N, Nakayama M, Eto H, Okumura K, Yagita H . Type I interferons (IFNs) regulate tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) expression on human T cells: a novel mechanism for the antitumor effects of type I IFNs. J Exp Med 1999; 189: 1451–1460.
Kaplan MJ, Ray D, Mo RR, Yung RL, Richardson BC . TRAIL (Apo2 ligand) and TWEAK (Apo3 ligand) mediate CD4+ T cell killing of antigen-presenting macrophages. J Immunol 2000; 164: 2897–2904.
Walczak H, Sprick MR . Biochemistry and function of the DISC. Trends Biochem Sci 2001; 26: 452–453.
Sprick MR, Weigand MA, Rieser E, Rauch CT, Juo P, Blenis J . FADD/MORT1 and caspase-8 are recruited to TRAIL receptors 1 and 2 and are essential for apoptosis mediated by TRAIL receptor 2. Immunity 2000; 12: 599–609.
Kischkel FC, Lawrence DA, Chuntharapai A, Schow P, Kim KJ, Ashkenazi A . Apo2L/TRAIL-dependent recruitment of endogenous FADD and caspase-8 to death receptors 4 and 5. Immunity 2000; 12: 611–620.
Bodmer JL, Holler N, Reynard S, Vinciguerra P, Schneider P, Juo P et al. TRAIL receptor-2 signals apoptosis through FADD and caspase-8. Nat Cell Biol 2000; 2: 241–243.
Sprick MR, Rieser E, Stahl H, Grosse-Wilde A, Weigand M A, Walczak H . Caspase-10 is recruited to and activated at the native TRAIL and CD95 death-inducing signalling complexes in a FADD-dependent manner but cannot functionally substitute caspase-8. EMBO J 2002; 21: 4520–4530.
Kischkel FC, Lawrence DA, Tinel A, LeBlanc H, Virmani A, Schow P et al. Death receptor recruitment of endogenous caspase-10 and apoptosis initiation in the absence of caspase-8. J Biol Chem 2001; 276: 46639–46646.
Wu YY, Tsai HF, Lin WC, Chou AH, Chen HT, Yang JC et al. Helicobacter pylori enhances tumor necrosis factor-related apoptosis-inducing ligand-mediated apoptosis in human gastric epithelial cells. World J Gastroenterol 2004; 10: 2334–2339.
Chou AH, Tsai HF, Wu YY, Hu CY, Hwang LH, Hsu PI et al. Hepatitis C virus core protein modulates TRAIL-mediated apoptosis by enhancing Bid cleavage and activation of mitochondria apoptosis signaling pathway. J Immunol 2005; 174: 2160–2166.
Nagane M, Pan G, Weddle JJ, Dixit VM, Cavenee WK, Huang HJ . Anticancer agents sensitize tumor cells to tumor necrosis factorrelated apoptosis-inducing ligand-mediated caspase-8 activation and apoptosis. Cancer Res 2000; 61: 1645–1651.
Wen J, Ramadevi N, Nguyen D, Perkins C, Worthington E, Bhalla K . Antileukemic drugs increase death receptor 5 levels and enhance Apo-2L-induced apoptosis of human acute leukemia cells. Blood 2000; 96: 3900–3906.
Belka C, Schmid B, Marini P, Durand E, Rudner J, Faltin H et al. Sensitization of resistant lymphoma cells to irradiation-induced apoptosis by the death ligand TRAIL. Oncogene 2001; 20: 2190–2196.
Ganten TM, Haas TL, Sykora J, Stahl H, Sprick MR, Fas SC et al. Enhanced caspase-8 recruitment to and activation at the DISC is critical for sensitisation of human hepatocellular carcinoma cells to TRAIL-induced apoptosis by chemotherapeutic drugs. Cell Death Differ 2004; 11: S86–S96.
Reers M, Smith TW, Chen LB . J-aggregate formation of a carbocyanine as a quantitative fluorescence indicator of membrane potential. Biochemistry 1991; 30: 448860–448844.
Smiley ST, Reers M, Mottola-Hartshorn C, Lin M, Chen A, Smith TW et al. Intracellular heterogeneity in mitochondrial membrane potentials revealed by a J-aggregate-forming lipophilic cation JC-1. Proc Natl Acad Sci USA 1991; 88: 3671–3675.
Oishi M, Iizumi Y, Taniguchi T, Goi W, Miki T, Sakai T . Apigenin sensitizes prostate cancer cells to Apo2L/TRAIL by targeting adenine nucleotide translocase-2. PLoS One 2013; 8: e55922.
Abou El, Naga RN, Azab SS, El-Demerdash E, Shaarawy S, El-Merzabani M, Ammar ES . Sensitization of TRAIL-induced apoptosis in human hepatocellular carcinoma HepG2 cells by phytochemicals. Life Sci 2013; 92: 555–561.
Wang G, Wang X, Yu H, Wei S, Williams N, Holmes DL et al. Small-molecule activation of the TRAIL receptor DR5 in human cancer cells. Nat Chem Biol 2013; 9: 84–89.
Kim EY, Yu JS, Yang M, Kim AK . Sub-toxic dose of apigenin sensitizes HepG2 cells to TRAIL through ERK-dependent up-regulation of TRAIL receptor DR5. Mol Cells 2013; 35: 32–40.
Surget S, Chiron D, Gomez-Bougie P, Descamps G, Ménoret E, Bataille R et al. Cell death via DR5, but not DR4, is regulated by p53 in myeloma cells. Cancer Res 2012; 72: 4562–4573.
Irmler M, Thome M, Hahne M, Schneider P, Hofmann K, Steiner V et al. Inhibition of death receptor signals by cellular FLIP. Nature 1997; 388: 190–195.
Golk A, Brenner D, Fritsch C, Krammer PH, Lavrik IN . c-FLIPR, a new regulator of death receptor-induced apoptosis. J Biol Chem 2005; 280: 14507–14513.
Scaffidi C, Schmitz I, Krammer PH, Peter ME . The role of c-FLIP in modulation of CD95-induced apoptosis. J Biol Chem 1999; 274: 1541–1548.
Krueger A, Schmitz I, Baumann S, Krammer PH, Kirchhoff S . Cellular flice-inhibitory protein splice variants inhibit different steps of caspase-8 activation at the CD95 death-inducing signaling complex. J Biol Chem 2001; 276: 20633–20640.
Karttunen R, Karttunen T, Ekre H-P T, MacDonald TT . Interferon gamma and interleukin 4 secreting cells in the gastric antrum in Helicobacter pylori positive and negative gastritis. Gut 1995; 36: 341–345.
Lindholm C, Quiding-Jalrbrink M, Lonroth H, Hamlet A, Svennerholm A-M . Local cytokine response in Helicobacter pylori-infected subjects. Infect Immun 1998; 66: 5964–5971.
Chou AH, Tsai HF, Lin LL, Hsieh SL, Hsu PI, Hsu PN . Enhanced proliferation and increased IFN-gamma production in T cells by signal transduced through TNF-related apoptosis-inducing ligand. J Immunol 2001; 167: 1347–1352.
Tsai HF, Lai JJ, Chou A, Wang TF, Wu CS, Hsu PN . Induction of costimulation of human CD4 T cells by tumor necrosis factor-related apoptosis-inducing ligand: possible role in T cell activation in systemic lupus erythematosus. Arthritis Rheum 2004; 50: 629–639.
Huang S, Tsai H, Tzeng H, Liao H, Hsu P . Lipid raft assembly and Lck recruitment in TRAIL costimulation mediates NF-κB activation and T cell proliferation. J Immunol 2011; 186: 931–939.
Gao LY, Kwaik YA . The modulation of host cell apoptosis by intracellular bacterial pathogens. Trends Microbiol 2000; 8: 306–313.
Odenbreit S, Puls J, Sedlmaier B, Gerland E, Fischer W, Haas R . Translocation of Helicobacter pylori CagA into gastric epithelial cells by type IV secretion. Science 2000; 287: 1497–1501.
Asahi BM, Azuma T, Ito S, Ito Y, Suto H, Nagai Y et al. Helicobacter pylori CagA protein can be tyrosine phosphorylated in gastric epithelial cells. J Exp Med 2000; 191: 593–602.
Stein M, Rappuoli R, Covacci A . Tyrosine phosphorylation of the Helicobacter pylori CagA antigen after cag-driven host cell translocation. Proc Natl Acad Sci USA 2000; 97: 1263–1268.
Higashi H, Tsutsumi R, Muto S, Sugiyama T, Azuma T, Asaka M et al. SHP-2 tyrosine phosphatase as an intracellular target of Helicobacter pylori CagA. Science 2002; 295: 683–686.
Acknowledgements
We thank Dr. N Sutkowski for critically reviewing the manuscript and Ms. W-I Tsai and W-H Wu for assistance with H. pylori culture. This work was supported by grants from the National Science Council, Taiwan (102-2320-B-038-040-MY3 and 102-2321-B-002-002), National Taiwan University Hospital (VN-9801), and China Medical University (CMU96-266, CMU97-299).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by A Oberst
Author contributions
PNH and HFT designed the research; WCL, HFT, and HJL performed the research; WCL, PIH, CHT, and ALC analyzed and interpreted the data; WCL and YYW performed the statistical analysis; and HFT and PNH wrote the manuscript.
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Lin, WC., Tsai, HF., Liao, HJ. et al. Helicobacter pylori sensitizes TNF-related apoptosis-inducing ligand (TRAIL)-mediated apoptosis in human gastric epithelial cells through regulation of FLIP. Cell Death Dis 5, e1109 (2014). https://doi.org/10.1038/cddis.2014.81
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2014.81
Keywords
This article is cited by
-
H. pylori infection confers resistance to apoptosis via Brd4-dependent BIRC3 eRNA synthesis
Cell Death & Disease (2020)
-
c-FLIP is a target of the E3 ligase deltex1 in gastric cancer
Cell Death & Disease (2018)
-
Helicobacter pylori-induced IL-33 modulates mast cell responses, benefits bacterial growth, and contributes to gastritis
Cell Death & Disease (2018)
-
Distinct roles of TNF-related apoptosis-inducing ligand (TRAIL) in viral and bacterial infections: from pathogenesis to pathogen clearance
Inflammation Research (2016)