Abstract
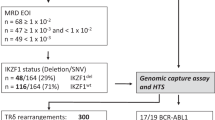
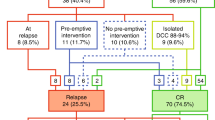
Minimal residual disease (MRD) monitoring via quantitative PCR (qPCR) detection of Ag receptor gene rearrangements has been the most sensitive method for predicting prognosis and making post-transplant treatment decisions for patients with ALL. Despite the broad clinical usefulness and standardization of this method, we and others have repeatedly reported the possibility of false-positive MRD results caused by massive B-lymphocyte regeneration after stem cell transplantation (SCT). Next-generation sequencing (NGS) enables precise and sensitive detection of multiple Ag receptor rearrangements, thus providing a more specific readout compared to qPCR. We investigated two cohorts of children with ALL who underwent SCT (30 patients and 228 samples). The first cohort consisted of 17 patients who remained in long-term CR after SCT despite having low MRD positivity (<0.01%) at least once during post-SCT monitoring using qPCR. Only one of 27 qPCR-positive samples was confirmed to be positive by NGS. Conversely, 10 of 15 samples with low qPCR-detected MRD positivity from 13 patients who subsequently relapsed were also confirmed to be positive by NGS (P=0.002). These data show that NGS has a better specificity in post-SCT ALL management and indicate that treatment interventions aimed at reverting impending relapse should not be based on qPCR only.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
van Dongen JJ, Seriu T, Panzer-Grümayer ER, Biondi A, Pongers-Willemse MJ, Corral L et al. Prognostic value of minimal residual disease in acute lymphoblastic leukaemia in childhood. Lancet (London, England) 1998; 352: 1731–1738.
van der Velden VHJ, Panzer-Grümayer ER, Cazzaniga G, Flohr T, Sutton R, Schrauder A et al. Optimization of PCR-based minimal residual disease diagnostics for childhood acute lymphoblastic leukemia in a multi-center setting. Leukemia 2007; 21: 706–713.
Fronkova E, Mejstrikova E, Avigad S, Chik KW, Castillo L, Manor S et al. Minimal residual disease (MRD) analysis in the non-MRD-based ALL IC-BFM 2002 protocol for childhood ALL: is it possible to avoid MRD testing? Leukemia 2008; 22: 989–997.
Pieters R, de Groot-Kruseman H, Van der Velden V, Fiocco M, van den Berg H, de Bont E et al. Successful therapy reduction and intensification for childhood acute lymphoblastic leukemia based on minimal residual disease monitoring: study ALL10 from the Dutch Childhood Oncology Group. J Clin Oncol 2016; 34: 2591–2601.
Knechtli CJ, Goulden NJ, Hancock JP, Grandage VL, Harris EL, Garland RJ et al. Minimal residual disease status before allogeneic bone marrow transplantation is an important determinant of successful outcome for children and adolescents with acute lymphoblastic leukemia. Blood 1998; 92: 4072–4079.
Krejci O, van der Velden VHJ, Bader P, Kreyenberg H, Goulden N, Hancock J et al. Level of minimal residual disease prior to haematopoietic stem cell transplantation predicts prognosis in paediatric patients with acute lymphoblastic leukaemia: a report of the Pre-BMT MRD Study Group. Bone Marrow Transplant 2003; 32: 849–851.
Sramkova L, Muzikova K, Fronkova E, Krejci O, Sedlacek P, Formankova R et al. Detectable minimal residual disease before allogeneic hematopoietic stem cell transplantation predicts extremely poor prognosis in children with acute lymphoblastic leukemia. Pediatr Blood Cancer 2007; 48: 93–100.
Spinelli O, Peruta B, Tosi M, Guerini V, Salvi A, Zanotti MC et al. Clearance of minimal residual disease after allogeneic stem cell transplantation and the prediction of the clinical outcome of adult patients with high-risk acute lymphoblastic leukemia. Haematologica 2007; 92: 612–618.
Bader P, Kreyenberg H, Henze GHR, Eckert C, Reising M, Willasch A et al. Prognostic value of minimal residual disease quantification before allogeneic stem-cell transplantation in relapsed childhood acute lymphoblastic leukemia: the ALL-REZ BFM Study Group. J Clin Oncol 2009; 27: 377–384.
Balduzzi A, Di Maio L, Silvestri D, Songia S, Bonanomi S, Rovelli A et al. Minimal residual disease before and after transplantation for childhood acute lymphoblastic leukaemia: is there any room for intervention? Br J Haematol 2014; 164: 396–408.
Bader P, Kreyenberg H, Von Stackelberg A, Eckert C, Salzmann-Manrique E, Meisel R et al. Monitoring of minimal residual disease after allogeneic stem-cell transplantation in relapsed childhood acute lymphoblastic leukemia allows for the identification of impending relapse: results of the all-bfm-sct 2003 trial. J Clin Oncol 2015; 33: 1275–1284.
Lankester AC, Bierings MB, van Wering ER, Wijkhuijs AJM, de Weger RA, Wijnen JT et al. Preemptive alloimmune intervention in high-risk pediatric acute lymphoblastic leukemia patients guided by minimal residual disease level before stem cell transplantation. Leukemia 2010; 24: 1462–1469.
van der Velden VHJ, van Dongen JJM . MRD detection in acute lymphoblastic leukemia patients using Ig/TCR gene rearrangements as targets for real-time quantitative PCR. Methods Mol Biol 2009; 538: 115–150.
van der Velden VHJ, Cazzaniga G, Schrauder A, Hancock J, Bader P, Panzer-Grumayer ER et al. Analysis of minimal residual disease by Ig/TCR gene rearrangements: guidelines for interpretation of real-time quantitative PCR data. Leukemia 2007; 21: 604–611.
van der Velden VHJ, Wijkhuijs JM, van Dongen JJM . Non-specific amplification of patient-specific Ig/TCR gene rearrangements depends on the time point during therapy: implications for minimal residual disease monitoring. Leukemia 2008; 22: 641–644.
Fronkova E, Muzikova K, Mejstrikova E, Kovac M, Formankova R, Sedlacek P et al. B-cell reconstitution after allogeneic SCT impairs minimal residual disease monitoring in children with ALL. Bone Marrow Transplant 2008; 42: 187–196.
Faham M, Zheng J, Moorhead M, Carlton VE, Stow P, Coustan-Smith E et al. Deep-sequencing approach for minimal residual disease detection in acute lymphoblastic leukemia. Blood 2012; 120: 5173–5180.
Kotrova M, Muzikova K, Mejstrikova E, Novakova M, Bakardjieva-Mihaylova V, Fiser K et al. The predictive strength of next-generation sequencing MRD detection for relapse compared with current methods in childhood ALL. Blood 2015; 126: 1045–1047.
Pulsipher MA, Carlson C, Langholz B, Wall DA, Schultz KR, Bunin N et al. IgH-V(D)J NGS-MRD measurement pre- and early post- allo-transplant defines very low and very high risk ALL patients. Blood 2015; 125: 3501–3508.
Pongers-Willemse MJ, Seriu T, Stolz F, d’Aniello E, Gameiro P, Pisa P et al. Primers and protocols for standardized detection of minimal residual disease in acute lymphoblastic leukemia using immunoglobulin and T cell receptor gene rearrangements and TAL1 deletions as PCR targets: report of the BIOMED-1 CONCERTED ACTION: investiga. Leukemia 1999; 13: 110–118.
van Dongen JJM, Langerak AW, Brüggemann M, Evans PAS, Hummel M, Lavender FL et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia 2003; 17: 2257–2317.
Langerak AW, Wolvers-Tettero IL, van Gastel-Mol EJ, Oud ME, van Dongen JJ . Basic helix-loop-helix proteins E2A and HEB induce immature T-cell receptor rearrangements in nonlymphoid cells. Blood 2001; 98: 2456–2465.
van der Velden VHJ, Wijkhuijs JM, Jacobs DCH, van Wering ER, van Dongen JJM . T cell receptor gamma gene rearrangements as targets for detection of minimal residual disease in acute lymphoblastic leukemia by real-time quantitative PCR analysis. Leukemia 2002; 16: 1372–1380.
van der Velden VHJ, Willemse MJ, van der Schoot CE, Hählen K, van Wering ER, van Dongen JJM . Immunoglobulin kappa deleting element rearrangements in precursor-B acute lymphoblastic leukemia are stable targets for detection of minimal residual disease by real-time quantitative PCR. Leukemia 2002; 16: 928–936.
Verhagen OJ, Willemse MJ, Breunis WB, Wijkhuijs AJ, Jacobs DC, Joosten SA et al. Application of germline IGH probes in real-time quantitative PCR for the detection of minimal residual disease in acute lymphoblastic leukemia. Leukemia 2000; 14: 1426–1435.
Pongers-Willemse MJ, Verhagen OJ, Tibbe GJ, Wijkhuijs AJ, de Haas V, Roovers E et al. Real-time quantitative PCR for the detection of minimal residual disease in acute lymphoblastic leukemia using junctional region specific TaqMan probes. Leukemia 1998; 12: 2006–2014.
Gabert J, Beillard E, van der Velden VHJ, Bi W, Grimwade D, Pallisgaard N et al. Standardization and quality control studies of ‘real-time’ quantitative reverse transcriptase polymerase chain reaction of fusion gene transcripts for residual disease detection in leukemia - a Europe Against Cancer program. Leukemia 2003; 17: 2318–2357.
Zaliova M, Fronkova E, Krejcikova K, Muzikova K, Mejstrikova E, Stary J et al. Quantification of fusion transcript reveals a subgroup with distinct biological properties and predicts relapse in BCR/ABL-positive ALL: implications for residual disease monitoring. Leukemia 2009; 23: 944–951.
Theunissen P, Mejstrikova E, Sedek L, van der Sluijs-Gelling AJ, Gaipa G, Bartels M et al. Standardized flow cytometry for highly sensitive MRD measurements in B-cell acute lymphoblastic leukemia. Blood 2016; 129: 347–357.
Szczepanski T, Pongers-willemse MJ, Langerak AW, Harts WA, Wijkhuijs AJM, Van Wering ER et al. Ig heavy chain gene rearrangements in T-cell acute lymphoblastic leukemia exhibit predominant DH6-19 and DH7-27 gene usage, can result in complete V-D-J rearrangements, and are rare in T-cell receptor alpha beta lineage. Blood 1999; 93: 4079–4085.
Brüggemann M, Schrauder A, Raff T, Pfeifer H, Dworzak M, Ottmann OG et al. Standardized MRD quantification in European ALL trials: proceedings of the Second International Symposium on MRD assessment in Kiel, Germany, 18-20 September 2008. Leukemia 2010; 24: 521–535.
Acknowledgements
This work was supported by 16-32568A (EF, MK and JT). JZ was supported by GBP302/12/G101 from Grant agency of the Czech Republic. MK was supported by GAUK 394214. JS was supported by institutional support University Hospital Motol 00064203 from Ministry of Health of the Czech Republic. We gratefully acknowledge Dr Arjan Lankester (LUMC, Leiden, the Netherlands) and Dr Marc Bierings (UMCU, Utrecht, the Netherlands) for providing clinical data for the Dutch patients and Mrs Rianne Noordijk, Mrs Maaike de Bie and Mrs Patricia Hoogeveen (Erasmus MC, Rotterdam, the Netherlands) for technical support.
Author contributions
MK prepared the sequencing libraries, performed the sequencing runs and performed the data analysis; VHJvdV, JJMvD, RF, PS, MB and JZ analyzed and interpreted the data; JS and JT planned the study and interpreted the data; and EF planned the study, interpreted the data and wrote the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
JJMvD, VHJvdV and MB are Board members of EuroMRD (www.EuroMRD.org). The remaining authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on Bone Marrow Transplantation website
Supplementary information
Rights and permissions
About this article
Cite this article
Kotrova, M., van der Velden, V., van Dongen, J. et al. Next-generation sequencing indicates false-positive MRD results and better predicts prognosis after SCT in patients with childhood ALL. Bone Marrow Transplant 52, 962–968 (2017). https://doi.org/10.1038/bmt.2017.16
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bmt.2017.16
This article is cited by
-
Clinical application of next-generation sequencing-based monitoring of minimal residual disease in childhood acute lymphoblastic leukemia
Journal of Cancer Research and Clinical Oncology (2023)
-
Predictive value of next-generation sequencing-based minimal residual disease after CAR-T cell therapy
Bone Marrow Transplantation (2022)
-
Application of Next-Generation Sequencing-Based Mutational Profiling in Acute Lymphoblastic Leukemia
Current Hematologic Malignancy Reports (2021)
-
Quality control and quantification in IG/TR next-generation sequencing marker identification: protocols and bioinformatic functionalities by EuroClonality-NGS
Leukemia (2019)
-
Standardized next-generation sequencing of immunoglobulin and T-cell receptor gene recombinations for MRD marker identification in acute lymphoblastic leukaemia; a EuroClonality-NGS validation study
Leukemia (2019)