Abstract
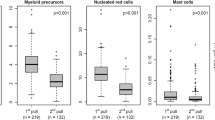
Enrichment of cell subpopulations is a prerequisite for lineage-specific chimerism analysis (LCA), a frequent approach in follow-up after allo-SCT. An efficient enrichment technique is Magnetic Cell Sorting (MACS) using the AutoMACS separator. However, evaluation of purity, recovery and applicability for PCR-based chimerism analysis of MACS-enriched subpopulations from post-transplant peripheral blood, providing reduced cell numbers and/or unbalanced proportions of subpopulations, is currently unavailable. We performed enrichment of CD3-, CD14-, CD15-, CD19- and CD56-positive subpopulations using ‘Whole Blood MicroBeads’ and AutoMACS separator in 137 prospectively collected peripheral blood samples from 15 paediatric patients after allo-CD3-/CD19-depleted SCT. Purity was assessed by immune phenotyping. Recovery and applicability for chimerism analysis was evaluated. Excellent purity >90% was achieved in CD14-, CD15-positive cells in 81%, 95% of the isolates and in 86% of CD3 and CD19 isolates, if ACC was >400 cells per μl. Median purity of CD56-positive isolates was 78.9%. Recovery >90% was between 93 (CD56) and 37% (CD15). Conventional and real-time PCR-based chimerism analysis was feasible in virtually all samples. Isolation of cell subpopulations by automated cell enrichment in post-transplant peripheral blood is feasible and fast providing excellent purity and recovery for routine lineage-specific chimerism analysis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Bader P, Kreyenberg H, Hoelle W, Dueckers G, Handgretinger R, Lang P et al. Increasing mixed chimerism is an important prognostic factor for unfavorable outcome in children with acute lymphoblastic leukemia after allogeneic stem-cell transplantation: possible role for pre-emptive immunotherapy? J Clin Oncol. 2004; 22: 1696–1706.
Willasch A, Hoelle W, Kreyenberg H, Niethammer D, Handgretinger R, Lang P et al. Outcome of allogeneic stem cell transplantation in children with non-malignant diseases. Haematologica 2006; 91: 788–794.
Thiede C, Bornhauser M, Ehninger G . Strategies and clinical implications of chimerism diagnostics after allogeneic hematopoietic stem cell transplantation. Acta Haematol 2004; 112: 16–23.
Liesveld JL, Rothberg PG . Mixed chimerism in SCT: conflict or peaceful coexistence? Bone Marrow Transplant 2008; 42: 297–310.
Saito B, Fukuda T, Yokoyama H, Kurosawa S, Takahashi T, Fuji S et al. Impact of T cell chimerism on clinical outcome in 117 patients who underwent allogeneic stem cell transplantation with a busulfan-containing reduced-intensity conditioning regimen. Biol Blood Marrow Transplant 2008; 14: 1148–1155.
Ozyurek E, Cowan MJ, Koerper MA, Baxter-Lowe LA, Dvorak CC, Horn BN . Increasing mixed chimerism and the risk of graft loss in children undergoing allogeneic hematopoietic stem cell transplantation for non-malignant disorders. Bone Marrow Transplant 2008; 42: 83–91.
Lion T, Muller-Berat N . Chimerism testing after allogeneic stem cell transplantation: importance of timing and optimal technique for testing in different clinical-biological situations. Leukemia 2003; 17: 612.
Matthes-Martin S, Lion T, Haas OA, Frommlet F, Daxberger H, Konig M et al. Lineage-specific chimaerism after stem cell transplantation in children following reduced intensity conditioning: potential predictive value of NK cell chimaerism for late graft rejection. Leukemia 2003; 17: 1934–1942.
Lim ZY, Pearce L, Ingram W, Ho AY, Mufti GJ, Pagliuca A . Chimerism does not predict for outcome after alemtuzumab-based conditioning: lineage-specific analysis of chimerism of specific diseases may be more informative. Bone Marrow Transplant 2008; 41: 587–588.
Ringden O . Immunotherapy by allogeneic stem cell transplantation. Adv Cancer Res 2007; 97C: 25–60.
Lion T . Detection of impending graft rejection and relapse by lineage-specific chimerism analysis. Methods Mol Med 2007; 134: 197–216.
Miura Y, Tanaka J, Toubai T, Tsutsumi Y, Kato N, Hirate D et al. Analysis of donor-type chimerism in lineage-specific cell populations after allogeneic myeloablative and non-myeloablative stem cell transplantation. Bone Marrow Transplant. 2006; 37: 837–843.
Zeiser R, Spyridonidis A, Wasch R, Ihorst G, Grullich C, Bertz H et al. Evaluation of immunomodulatory treatment based on conventional and lineage-specific chimerism analysis in patients with myeloid malignancies after myeloablative allogeneic hematopoietic cell transplantation. Leukemia 2005; 19: 814–821.
Bader P, Stoll K, Huber S, Geiselhart A, Handgretinger R, Niemeyer C et al. Characterization of lineage-specific chimaerism in patients with acute leukaemia and myelodysplastic syndrome after allogeneic stem cell transplantation before and after relapse. Br J Haematol 2000; 108: 761–768.
Breuer S, Matthes S, König M, Fritsch G, Pötschger U, Lawitschka A et al. Recipient chimerism in the T-cell lineage is indicative of impending graft rejection in pediatric patients undergoing allogeneic hematopoietic stem cell transplantation. In: 40th Congress of SIOP, abstract book. Wiley-Liss Inc.: Hoboken, NY, USA, 2008, pp 60–61.
Pongers-Willemse MJ, Verhagen OJ, Tibbe GJ, Wijkhuijs AJ, de Haas V, Roovers E et al. Real-time quantitative PCR for the detection of minimal residual disease in acute lymphoblastic leukemia using junctional region specific TaqMan probes. Leukemia 1998; 12: 2006–2014.
Kreyenberg H, Holle W, Mohrle S, Niethammer D, Bader P . Quantitative analysis of chimerism after allogeneic stem cell transplantation by PCR amplification of microsatellite markers and capillary electrophoresis with fluorescence detection: the Tuebingen experience. Leukemia 2003; 17: 237–240.
Watzinger F, Lion T, Steward C . The RSD code: proposal for a nomenclature of allelic configurations in STR-PCR-based chimerism testing after allogeneic stem cell transplantation. Leukemia 2006; 20: 1448–1452.
Scharf SJ, Smith AG, Hansen JA, McFarland C, Erlich HA . Quantitative determination of bone marrow transplant engraftment using fluorescent polymerase chain reaction primers for human identity markers. Blood 1995; 85: 1954–1963.
Willasch A, Schneider G, Reincke BS, Shayegi N, Kreyenberg H, Kuci S et al. Sequence polymorphism systems for quantitative real-time polymerase chain reaction to characterize hematopoietic chimerism-high informativity and sensitivity as well as excellent reproducibility and precision of measurement. Lab Hematol 2007; 13: 73–84.
Steinbach D, Debatin KM . What do we mean by sensitivity when we talk about detecting minimal residual disease? Leukemia 2008; 22: 1638–1639.
The R Foundation for Statistical Computing c/o Institut für Statistik und Wahrscheinlichkeitstheorie der Technischen Universität Wien. R, a language and environment for statistical computing and graphics. University of Vienna, Austria 2003.
Vitale M, Della CM, Carlomagno S, Romagnani C, Thiel A, Moretta L . et al. The small subset of CD56brightCD16- natural killer cells is selectively responsible for both cell proliferation and interferon-gamma production upon interaction with dendritic cells. Eur J Immunol 2004; 34: 1715–1722.
Romagnani C, Juelke K, Falco M, Morandi B, D'Agostino A, Costa R et al. CD56brightCD16- killer Ig-like receptor- NK cells display longer telomeres and acquire features of CD56dim NK cells upon activation. J Immunol 2007; 178: 4947–4955.
Beck O, Seidl C, Lehrnbecher T, Kreyenberg H, Schwabe D, Klingebiel T et al. Quantification of chimerism within peripheral blood, bone marrow and purified leukocyte subsets: comparison of singleplex and multiplex PCR amplification of short tandem repeat (STR) loci. Eur J Haematol. 2006; 76: 237–244.
Tomblyn M, Lazarus HM . Donor lymphocyte infusions: the long and winding road: how should it be traveled? Bone Marrow Transplant 2008.
Klingebiel T, Bader P . Delayed lymphocyte infusion in children given SCT. Bone Marrow Transplant 2008; 41 (Suppl 2): S23–S26.
Formankova R, Sedlacek P, Krskova L, Rihova H, Sramkova L, Star J . Chimerism-directed adoptive immunotherapy in prevention and treatment of post-transplant relapse of leukemia in childhood. Haematologica 2003; 88: 117–118.
Acknowledgements
This work was supported by the ‘Adolf Messer Stiftung’, Koenigstein, Germany (PB) and by the ‘Paul und Ursula Klein-Stiftung’, Frankfurt, Germany (AW).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Willasch, A., Eing, S., Weber, G. et al. Enrichment of cell subpopulations applying automated MACS technique: purity, recovery and applicability for PCR-based chimerism analysis. Bone Marrow Transplant 45, 181–189 (2010). https://doi.org/10.1038/bmt.2009.89
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bmt.2009.89
Keywords
This article is cited by
-
Gene expression profiling of immunomagnetically separated cells directly from stabilized whole blood for multicenter clinical trials
Clinical and Translational Medicine (2014)