Abstract
Background:
Prostate-specific antigen (PSA) and PSA-velocity (PSAV) have been used to identify men at risk of prostate cancer (PrCa). The IMPACT study is evaluating PSA screening in men with a known genetic predisposition to PrCa due to BRCA1/2 mutations. This analysis evaluates the utility of PSA and PSAV for identifying PrCa and high-grade disease in this cohort.
Methods:
PSAV was calculated using logistic regression to determine if PSA or PSAV predicted the result of prostate biopsy (PB) in men with elevated PSA values. Cox regression was used to determine whether PSA or PSAV predicted PSA elevation in men with low PSAs. Interaction terms were included in the models to determine whether BRCA status influenced the predictiveness of PSA or PSAV.
Results:
1634 participants had ⩾3 PSA readings of whom 174 underwent PB and 45 PrCas diagnosed. In men with PSA >3.0 ng ml−l, PSAV was not significantly associated with presence of cancer or high-grade disease. PSAV did not add to PSA for predicting time to an elevated PSA. When comparing BRCA1/2 carriers to non-carriers, we found a significant interaction between BRCA status and last PSA before biopsy (P=0.031) and BRCA2 status and PSAV (P=0.024). However, PSAV was not predictive of biopsy outcome in BRCA2 carriers.
Conclusions:
PSA is more strongly predictive of PrCa in BRCA carriers than non-carriers. We did not find evidence that PSAV aids decision-making for BRCA carriers over absolute PSA value alone.
Similar content being viewed by others
Main
Men with germline mutations in BRCA2 have an increased risk of prostate cancer (PrCa), estimated at 2.5–8.6 fold increased risk for BRCA2 mutation carriers (Breast Cancer Linkage Consortium, 1999; van Asperen et al, 2005; Kote-Jarai et al, 2011). There remains debate about whether there is an increased risk of PrCa associated with BRCA1 mutations, with some studies reporting no increased risk to those reporting a 1.8–3.75 fold increased risk (Thompson et al, 2002; Leongamornlert et al, 2012; Moran et al, 2012). A number of studies have reported that BRCA2 mutation carriers have more aggressive disease, suggested by their younger age at diagnosis, higher rates of lymph node involvement and distant metastasis at diagnosis, and higher mortality rates compared with non-carriers (Tryggvadóttir et al, 2007; Mitra et al, 2008; Edwards et al, 2010; Gallagher et al, 2010; Thorne et al, 2011; Castro et al, 2013). There is increasing evidence that BRCA1 mutation carriers may also harbour more aggressive disease (Giusti et al, 2003; Gallagher et al, 2010; Castro et al, 2013). Furthermore, BRCA2-mutant localised prostate cancer demonstrates increased genomic instability and a mutational profile that more closely resembles metastastic than localised disease, therefore supporting early detection in this at risk patient population (Taylor et al, 2017).
General population prostate specific antigen (PSA) screening remains controversial due to an unclear balance of benefits, in terms of mortality reduction when compared to harms such as overdiagnosis and overtreatment. However, many expert groups continue to recommend PrCa screening with particular attention towards men with risk factors based on family history, genetics and/or race (Roobol et al, 2013; Eeles et al, 2014; Mikropoulos et al, 2014; Murphy et al, 2014).
It has previously been suggested that the rate of PSA change over time, or PSA velocity (PSAV), can be used to assist in differentiating between men with cancer from those with benign disease (Carter et al, 1992; Berger et al, 2005). Monitoring PSA over time could also improve the sensitivity of screening. It is also possible that PSAV could distinguish between men who might have advanced or aggressive disease that would require definitive treatment thus avoiding overdiagnosis and overtreatment (Carter et al, 2006; Carter et al, 2007).
However, the utility of PSAV in PrCa decision-making has been called into question. In particular, while PSAV may be predictive of biopsy outcome in univariate analyses, it has not been shown to improve the predictiveness of biopsy outcome over the absolute value of PSA (Roobol et al, 2004; Vickers et al, 2011; Loughlin, 2014). Although some studies have suggested that PSAV can be used to identify men with aggressive disease, these did not investigate whether calculation of PSAV provided additional information than the most recent PSA value (Carter et al, 1992; D'Amico et al, 2004; D'Amico et al, 2005). PSA and PSAV are highly correlated, and this may explain why PSAV does not add predictive value (Vickers et al, 2011). As a result of these considerations, PSAV has been removed from all major guidelines concerning the detection of prostate cancer.
It is currently unknown whether PSAV provides more information, beyond PSA absolute value, among a cohort of men considered to be at increased genetic risk of PrCa and aggressive disease. The IMPACT study (Identification of Men with a genetic predisposition to ProstAte Cancer: Targeted screening in men at higher genetic risk and controls; www.impact-study.co.uk) is an international multi-centre study evaluating the role of targeted PSA screening in men with a BRCA1 or BRCA2 mutation and was established in 2005 (Bancroft et al, 2014). To date, ∼3000 men have been recruited from 20 countries across the world. Men are followed up with annual (or biannual in the Dutch cohort) PSA screening for a minimum of 5 years within the study and this has produced a wealth of PSA results and follow-up data over time. The primary end-point of the IMPACT study is to determine the incidence, stage and pathology of screen-detected prostate cancer in the study population; a secondary end-point is to determine a profile of PSA level and its predictive value for the development of prostate cancer in the study population. The objective of the present study was to determine whether PSA values and/or PSAV were associated with PrCa and aggressive tumours among men at increased risk enrolled in the IMPACT trial.
Materials and methods
Patient selection
The design and eligibility criteria for the IMPACT study have been described elsewhere (Mitra et al, 2008; Bancroft et al, 2014). The protocol was approved by the West-Midlands Research and Ethics Committee in the UK (reference 05/MRE07/25), and subsequently by each participating institution’s local committee. Briefly, men aged between 40 and 69 were recruited from families with a known pathogenic germline BRCA1 or BRCA2 mutation. Men were invited to enrol if they had tested positive (carriers) or negative for the familial mutation (BRCA1/2 non-carriers), or if they were at 50% risk of inheriting a mutation but had not yet undergone predictive genetic testing. All participants provide written consent. Men with PrCa or with a prior diagnosis of another cancer with a prognosis of <5 years were excluded. In the Dutch centres, men were also excluded if they had PSA screening prior to study entry.
According to the IMPACT study design, men underwent annual PSA screening and those with a PSA >3.0 ng ml−l were referred for a 10- or 12-core transrectal ultrasound guided (TRUS) biopsy based on institutional clinic practices. Men with a PSA >3.0 ng/ml and a negative biopsy continue annual screening, with a repeat biopsy recommended when PSA increased by >50%. Men were also referred for biopsy if they had a PSA ⩽3.0 ng ml−l but clinical suspicion (e.g., abnormal digital rectal examination or clinical symptoms). After 5 years in the study, men at a subset of centres were also offered an elective biopsy.
PSA readings in the study are validated in a central laboratory to exclude inter-site variations. The results found a Spearman’s agreement of 0.95 between study sites (Bancroft et al, 2014).
Statistical considerations
PSA velocity (PSAV) has been used as a marker to inform decisions about biopsy or about the timing of the next PSA screen. With respect to the former, we considered that a physician had the most recent PSA measurements available. Our study question was therefore whether adding PSAV to this data point improves prediction of presence of PrCa at biopsy. As elevated PSA is the primary indication in routine clinical practice, our main analysis was restricted to men who had any PSA ⩾3.0 ng ml−l prior to biopsy. A sensitivity analysis was conducted including all men who underwent biopsy. We created logistic regression models, adjusted for last PSA measurement and age, for the outcomes of any grade and high-grade cancer. PSAV was calculated using three methods: arithmetic equation of change in PSA over time; linear regression; rate of PSA change using first and last values only. We also used cubic splines with knots at the tertiles to test for non-linearity in PSA and in PSAV.
To investigate whether the effect of PSAV on predicting biopsy outcome differed based on BRCA status, we included an interaction term between PSAV and BRCA status (BRCA1 or BRCA2 carriers vs BRCA non-carrier patients, and BRCA2 carriers vs BRCA1 and BRCA non-carrier patients). Due to a limited number of events, this analysis was performed only for the outcome of any cancer on biopsy. This analysis included 13 cancers diagnosed among 55 BRCA1 carriers and 23 cancers among 65 BRCA2 carriers.
To determine whether PSAV could aid decisions about screening frequency, e.g., whether a man with a high PSAV should receive a subsequent PSA test at a shorter interval than a man with low PSAV, we assessed whether PSAV was associated with having a future PSA >3.0 ng ml−l. As a minimum of three PSA measurements are required for accurate estimation of PSAV, we created Cox proportional hazards models for the time from the first PSA measurement to the patient’s third PSA measurement >3.0 ng ml−l. Four models were then created: one including the third PSA measurement only, and the others including both the third PSA measurement and each of the three methodologies for calculating PSAV. Men who had a PSA >3.0 ng ml−l within the first three PSA measurements were excluded from this analysis. A total of 1086 men were included.
We planned to first evaluate the independent statistical significance of PSAV in models that also included absolute PSA level. If significant, we planned to estimate the improvement in concordance index afforded by PSAV after 10-fold cross-validation. All analyses were conducted using Stata 13.0 (Stata Corp., College Station, TX, USA).
Results
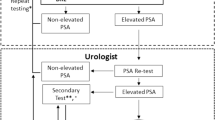
Of the 2942 men recruited to the IMPACT study, 1654 men had three or more PSA measurements and appropriate clinical follow-up to be included in the analyses (Figure 1). Table 1 shows the demographic, PSA and biopsy grade characteristics of the analysis cohorts. The cohort of 1654 men consisted of 510 BRCA1 mutation carriers, 584 BRCA2 mutation carriers, 260 BRCA1 non-carriers and 288 BRCA2 non-carriers. Two men carried both a BRCA1 and BRCA2 mutation (included in the BRCA2 group for genetic sub-analysis) and 10 had not yet had a predictive test for the BRCA mutation in their family (excluded from genetic sub-analysis). In this cohort, 174 men underwent prostate biopsy, with 45 men having any grade cancer of whom 21 having Gleason score 7 or higher (high-grade) cancer. Among men who had any PSA >3.0 ng ml−l, 40 had any grade of whom 20 had high-grade cancer.
The median age at the first PSA of BRCA2 carriers was significantly younger than both BRCA1 carriers and non-carriers (51 vs 53 vs 54 years, respectively, P<0.0001). Overall, BRCA2 and BRCA1 carriers had significantly lower first PSA values than non-carriers (0.80 vs 0.80 vs 0.89 ng ml−l; P=0.022; however, overall there was no statistically significant difference in the median PSAV between the BRCA2, BRCA1 and non-carrier groups (P=0.8).
The median age at first PSA reading of men diagnosed with cancer was higher than that of men without cancer (60 vs 53 years, U=22069, z=−5.24, P<0.001). The median most recent PSA (i.e., PSA at diagnosis for cancer cases) was significantly higher for those with cancer compared with those without cancer (3.70 vs 0.90 ng ml−l) U=8564, z=−9.34, P<0.001). The median PSAV was significantly higher for those with cancer vs those without cancer (medians: 0.56 vs 0.02 ng/ml/yr, U=9641, z=−9.012, P<0.001). Of those diagnosed with cancer, there was no significant difference between the proportion of BRCA2 carriers with a PSAV (calculated by linear regression) >0.35 ng ml−l per year compared with BRCA1 carriers and non-carrier controls (78.3 vs 61.5 vs 53.8%, p0.28).
We next assessed whether adding PSAV to the most recent PSA measurement would improve the ability to determine which men should undergo biopsy. Using cubic splines, we investigated and found no evidence of non-linearity in PSA or in PSAV. Among men with any PSA measurements >3.0 ng/ml, PSAV was not significantly associated with either any grade or high-grade cancer after adjusting for most recent PSA measurement (Table 2). We repeated these analyses including all men who were biopsied, and found that PSAV was not statistically significant in any of the models (Table 3).
Additionally, we assessed whether PSAV affected the prediction of PrCa at biopsy differently based on BRCA status. When comparing BRCA1 and BRCA2 carriers to BRCA1/2 non-carriers, we found a significant interaction between BRCA status and the last PSA before biopsy (P=0.031), however there was no evidence of an interaction between BRCA status and PSAV (Table 4). However, when comparing BRCA2 carriers to BRCA1 carriers and BRCA1/2 non-carriers, we found evidence of interactions between BRCA2 status and last PSA before biopsy (P=0.078) and significant interactions between BRCA2 status and PSAV calculated using the arithmetic equation and linear regression (P=0.024 and P=0.049 respectively, Table 4).
Based on these interactions, we performed subgroup analyses by BRCA2 status. All models were adjusted for age at biopsy and last PSA before biopsy. Due to a limited number of events (26 cancers in BRCA2 non-carriers and 23 in BRCA2 carriers), these models were somewhat overfit. No evidence of an association between PSA and any grade cancer or PSAV and any grade cancer was seen in BRCA2 carriers or non-carriers, likely due to the strong correlation between PSA and PSAV (Table 5).
We then investigated whether PSAV was associated with time to PSA ⩾3.0 ng ml−l. Using Cox proportional hazards models, we found no evidence of an association between PSAV and time from the third PSA measurement to PSA ⩾3.0 ng ml−l. Out of 1533 men who did not have a PSA ⩾3.0 ng ml−l within the first three PSA tests, there were 28 who had a PSA ⩾3.0 ng ml−l within 1 year, 50 within 2 years and 62 within 3 years.
Discussion
This is the first study to show that there are differences in PSA values among men with different genetic backgrounds. These PSA differences could be used to identify those men considered to be at high genetic risk of more aggressive disease. However, when evaluated with absolute PSA values, PSAV did not appear to provide additional information for BRCA1 or BRCA2 carriers.
A major problem of PSA screening is that, in attempting to detect clinically significant disease, it is inevitable that indolent disease will also be detected leading to overdiagnosis. However, early diagnosis and identification of men with high-risk disease is important to prevent mortality from PrCa. This might be particularly essential in light of recent publications indicating that men with a BRCA1 or BRCA2 mutation are at risk of more aggressive disease (Castro et al, 2013; Castro et al, 2015), early identification of those with clinically significant disease will be imperative. In view of the controversy about the role of PSAV in prostate screening in the general population, it was important to assess its role in BRCA1 and BRCA2 carriers and whether it added to the ability to detect clinically significant disease.
In this analysis of the IMPACT study cohort, we found BRCA2 carriers on average to be screened at a relatively young age. This may account for lower overall PSA values for BRCA2 carriers in this analysis compared to non-carrier controls. However, there were no differences in median PSAV between carriers and non-carriers. Given the possibility that higher PSAV may associate with aggressive PrCa (D'Amico et al, 2004; D'Amico et al, 2005), we would expect BRCA2 carriers who are at risk of aggressive disease would exhibit higher PSAVs (Castro et al, 2013; Castro et al, 2015). BRCA2 carriers in this group may be too young to demonstrate this trend at this point of follow up.
A single PSA reading over 3 ng ml−l was applied to guide biopsy decisions according to the IMPACT protocol, as well as if there was clinical suspicion on digital rectal examination or clinical symptoms. PSAV was not a good indicator in this analysis for distinguishing between those with any grade cancer and high-grade cancer when men were biopsied for either indication. It is possible that PSAV could be a good predictor of high-grade disease in men who had PSA values ⩽2 ng ml−l (Kitagawa et al, 2014). However, due to the protocol’s 3 ng/ml PSA threshold for prostate biopsy, we were limited in the number of cancers diagnosed when PSA was ⩽2 ng ml−l. Further follow up will be required to assess when additional cancers are diagnosed. As part of the IMPACT trial, there is an optional end of study biopsy regardless of PSA. This may help delineate PSAV among men diagnosed with PrCa with low PSA values (Carter et al, 1992; Kitagawa et al, 2014).
A strength of this study is the unique patient cohort of men with a genetic predisposition to PrCa, in particular BRCA2 carriers who are predisposed to aggressive PrCas (Mitra et al, 2008; Narod et al, 2008; Castro et al, 2013; Castro et al, 2015). Within the group of BRCA2 carriers, PSAV proved predictive of any grade cancer, however, given the low number of cancers diagnosed overall it was not possible to assess whether PSAV was associated with high grade cancer and BRCA2 status. A high PSAV in an individual with a BRCA2 mutation could be used as an indicator of presence of PrCa and therefore as an indication for prostate biopsy. This model could lead to diagnosis of lower grade cancers in BRCA2 carriers. It may lead to better prognosis for men at risk for more aggressive disease and better disease-free survival when treated early. Although there are no definitive treatment recommendations for men with BRCA2 mutations when found to be diagnosed with low grade cancers, their risk for aggressive disease may spur them to follow a more active treatment plan such as radical prostatectomy vs external-beam radiation therapy or active surveillance (Bratt and Loman, 2015; Castro et al, 2015).
One major limitation of this analysis is the relatively small number of men in the study who had undergone diagnostic prostate biopsy. End of study biopsies are not mandated and the true incidence of PrCa is unknown in this population. As more men progress through the IMPACT screening study, undergo prostate biopsy and follow-up time increases, the findings from this analysis can be explored and validated. At this point, the results from this analysis did not justify modifying the study algorithm to include a PSAV calculation.
In the general population, PSAV is not part of any major screening guideline. We also did not find PSAV to be an independent prognostic factor in BRCA1 or BRCA2 mutation carriers and therefore for screening an absolute PSA cut-off value should preferably be used.
Conclusion
PSA is more strongly predictive of PrCa in BRCA carriers than BRCA non-carriers. We did not find evidence that PSAV aids to decision making for either indicating biopsy or frequency of follow-up testing in BRCA carriers, but further follow-up is required for more definitive conclusions.
References
Bancroft EK, Page EC, Castro E, Lilja H, Vickers A, Sjoberg D, Assel M, Foster CS, Mitchell G, Drew K, Mæhle L, Axcrona K, Evans DG, Bulman B, Eccles D, McBride D, van Asperen C, Vasen H, Kiemeney LA, Ringelberg J, Cybulski C, Wokolorczyk D, Selkirk C, Hulick PJ, Bojesen A, Skytte AB, Lam J, Taylor L, Oldenburg R, Cremers R, Verhaegh G, van Zelst-Stams WA, Oosterwijk JC, Blanco I, Salinas M, Cook J, Rosario DJ, Buys S, Conner T, Ausems MG, Ong KR, Hoffman J, Domchek S, Powers J, Teixeira MR, Maia S, Foulkes WD, Taherian N, Ruijs M, Helderman-van den Enden AT, Izatt L, Davidson R, Adank MA, Walker L, Schmutzler R, Tucker K, Kirk J, Hodgson S, Harris M, Douglas F, Lindeman GJ, Zgajnar J, Tischkowitz M, Clowes VE, Susman R, Ramón y Cajal T, Patcher N, Gadea N, Spigelman A, van Os T, Liljegren A, Side L, Brewer C, Brady AF, Donaldson A, Stefansdottir V, Friedman E, Chen-Shtoyerman R, Amor DJ, Copakova L, Barwell J, Giri VN, Murthy V, Nicolai N, Teo SH, Greenhalgh L, Strom S, Henderson A, McGrath J, Gallagher D, Aaronson N, Ardern-Jones A, Bangma C, Dearnaley D, Costello P, Eyfjord J, Rothwell J, Falconer A, Gronberg H, Hamdy FC, Johannsson O, Khoo V, Kote-Jarai Z, Lubinski J, Axcrona U, Melia J, McKinley J, Mitra AV, Moynihan C, Rennert G, Suri M, Wilson P, Killick E IMPACT Collaborators Moss S, Eeles RA (2014) Targeted prostate cancer screening in BRCA1 and BRCA2 mutation carriers: results from the initial screening round of the IMPACT Study. Eur Urol 66: 489–499.
Berger AP, Deibl M, Steiner H, Bektic J, Pelzer A, Spranger R, Klocker H, Bartsch G, Horninger W (2005) Longitudinal PSA changes in men with and without prostate cancer: assessment of prostate cancer risk. Prostate 64: 240–245.
Bratt O, Loman N (2015) Clinical management of prostate cancer in men with BRCA mutations. Eur Urol 68: 194–195.
Breast Cancer Linkage Consortium (1999) Cancer risks in BRCA2 mutation carriers. J Natl Cancer Inst 91: 1310–1316.
Carter HB, Pearson JD, Metter EJ, Brant LJ, Chan DW, Andres R, Fozard JL, Walsh PC (1992) Longitudinal evaluation of prostate-specific antigen levels in men with and without prostate disease. JAMA 267: 2215–2220.
Carter HB, Kettermann A, Ferrucci L, Kettermann A, Landis P, Wright EJ, Epstein JI, Trock BJ, Metter EJ (2006) Detection of life-threatening prostate cancer with prostate-specific antigen velocity during a window of curability. J Natl Cancer Inst 98: 1521–1527.
Carter HB, Kettermann A, Ferrucci L, Landis P, Metter EJ (2007) Prostate-specific antigen velocity risk count assessment: a new concept for detection of life-threatening prostate cancer during window of curability. Urol 70: 685–690.
Castro E, Goh C, Olmos D, Saunders E, Leongamornlert D, Tymrakiewicz M, Mahmud N, Dadaev T, Govindasami K, Guy M, Sawyer E, Wilkinson R, Ardern-Jones A, Ellis S, Frost D, Peock S, Evans DG, Tischkowitz M, Cole T, Davidson R, Eccles D, Brewer C, Douglas F, Porteous ME, Donaldson A, Dorkins H, Izatt L, Cook J, Hodgson S, Kennedy MJ, Side LE, Eason J, Murray A, Antoniou AC, Easton DF, Kote-Jarai Z, Eeles R (2013) Germline BRCA mutations are associated with higher risk of nodal involvement, distant metastasis, and poor survival outcomes in prostate cancer. J Clin Oncol 31: 1748–1757.
Castro E, Goh C, Leongamornlert D, Saunders E, Tymrakiewicz M, Dadaev T, Govindasami K, Guy M, Ellis S, Frost D, Bancroft E, Cole T, Tischkowitz M, Kennedy MJ, Eason J, Brewer C, Evans DG, Davidson R, Eccles D, Porteous ME, Douglas F, Adlard J, Donaldson A, Antoniou AC, Kote-Jarai Z, Easton DF, Olmos D, Eeles R (2015) Effect of BRCA mutations on metastatic relapse and cause-specific survival after radical treatment for localised prostate cancer. Eur Urol 68: 186–193.
D'Amico AV, Chen MH, Roehl KA, Catalona WJ (2004) Preoperative PSA velocity and the risk of death from prostate cancer after radical prostatectomy. N Engl J Med 351: 125–135.
D'Amico AV, Renshaw AA, Sussman B, Chen MH (2005) Pretreatment PSA velocity and risk of death from prostate cancer following external beam radiation therapy. JAMA 294: 440–447.
Edwards SM, Evans DG, Hope Q, Norman AR, Barbachano Y, Bullock S, Kote-Jarai Z, Meitz J, Falconer A, Osin P, Fisher C, Guy M, Jhavar SG, Hall AL, O'Brien LT, Gehr-Swain BN, Wilkinson RA, Forrest MS, Dearnaley DP, Ardern-Jones AT, Page EC, Easton DF, Eeles RA UK Genetic Prostate Cancer Study Collaborators and BAUS Section of Oncology (2010) Prostate cancer in BRCA2 germline mutation carriers is associated with poorer prognosis. Br J Cancer 103: 918–924.
Eeles R, Goh C, Castro E, Bancroft E, Guy M, Al Olama AA, Easton D, Kote-Jarai Z (2014) The genetic epidemiology of prostate cancer and its clinical implications. Nat Rev Urol 11: 18–31.
Gallagher DJ, Gaudet MM, Pal P, Kirchhoff T, Balistreri L, Vora K, Bhatia J, Stadler Z, Fine SW, Reuter V, Zelefsky M, Morris MJ, Scher HI, Klein RJ, Norton L, Eastham JA, Scardino PT, Robson ME, Offit K (2010) Germline BRCA mutations denote a clinicopathologic subset of prostate cancer. Clin Cancer Res 16: 2115–2121.
Giusti RM, Rutter JL, Duray PH, Freedman LS, Konichezky M, Fisher-Fischbein J, Greene MH, Maslansky B, Fischbein A, Gruber SB, Rennert G, Ronchetti RD, Hewitt SM, Struewing JP, Iscovich J (2003) A twofold increase in BRCA mutation related prostate cancer among Ashkenazi Israelis is not associated with distinctive histopathology. J Med Genet 40: 787–792.
Kitagawa Y, Sawada K, Urata S, Izumi K, Ueno S, Kadono Y, Konaka H, Mizokami A, Namiki M (2014) Impact of PSA levels on second-round screening for the development of prostate cancer in men with low baseline PSA levels (</=2.0 mg/ml). Anticancer Res 34: 6739–6746.
Kote-Jarai Z, Leongamornlert D, Saunders E, Tymrakiewicz M, Castro E, Mahmud N, Guy M, Edwards S, O'Brien L, Sawyer E, Hall A, Wilkinson R, Dadaev T, Goh C, Easton D UKGPCS Collaborators Goldgar D, Eeles R (2011) BRCA2 is a moderate penetrance gene contributing to young-onset prostate cancer: implications for genetic testing in prostate cancer patients. Br J Cancer 105: 1230–1234.
Leongamornlert D, Mahmud N, Tymrakiewicz M, Saunders E, Dadaev T, Castro E, Goh C, Govindasami K, Guy M, O'Brien L, Sawyer E, Hall A, Wilkinson R, Easton D UKGPCS Collaborators Goldgar D, Eeles R, Kote-Jarai Z (2012) Germline BRCA1 mutations increase prostate cancer risk. Br J Cancer 106: 1697–1701.
Loughlin KR (2014) PSA velocity: A systematic review of clinical applications. Urol Oncol 32 (8): 1116–1125.
Mikropoulos C, Goh C, Leongamornlert D, Kote-Jarai Z, Eeles R (2014) Translating genetic risk factors for prostate cancer to the clinic: 2013 and beyond. Future Oncol 10: 1679–1694.
Mitra A, Fisher C, Foster CS, Jameson C, Barbachanno Y, Bartlett J, Bancroft E, Doherty R, Kote-Jarai Z, Peock S, Easton D IMPACT and EMBRACE Collaborators Eeles R (2008) Prostate cancer in male BRCA1 and BRCA2 mutation carriers has a more aggressive phenotype. Br J Cancer 98: 502–507.
Moran A, O'Hara C, Khan S, Shack L, Woodward E, Maher ER, Lalloo F, Evans DG (2012) Risk of cancer other than breast or ovarian in individuals with BRCA1 and BRCA2 mutations. Fam Cancer 11 (2): 235–242.
Murphy DG, Ahlering T, Catalona WJ, Crowe H, Crowe J, Clarke N, Cooperberg M, Gillatt D, Gleave M, Loeb S, Roobol M, Sartor O, Pickles T, Wootten A, Walsh PC, Costello AJ (2014) The Melbourne Consensus Statement on the early detection of prostate cancer. BJU Int 113 (2): 186–188.
Narod SA, Neuhausen S, Vichodez G, Armel S, Lynch HT, Ghadirian P, Cummings S, Olopade O, Stoppa-Lyonnet D, Couch F, Wagner T, Warner E, Foulkes WD, Saal H, Weitzel J, Tulman A, Poll A, Nam R, Sun P Hereditary Breast Cancer Study Group Danquah J, Domchek S, Tung N, Ainsworth P, Horsman D, Kim-Sing C, Maugard C, Eisen A, Daly M, McKinnon W, Wood M, Isaacs C, Gilchrist D, Karlan B, Nedelcu R, Meschino W, Garber J, Pasini B, Manoukian S, Bellati C (2008) Rapid progression of prostate cancer in men with a BRCA2 mutation. Br J Cancer 99: 371–374.
Roobol MJ, Kranse R, de Koning HJ, Schroder FH (2004) Prostate-specific antigen velocity at low prostate-specific antigen levels as screening tool for prostate cancer: results of second screening round of ERSPC (ROTTERDAM). Urology 63: 309–313, discussion 313–315.
Roobol MJ, Kranse R, Bangma CH, van Leenders AG, Blijenberg BG, van Schaik RH, Kirkels WJ, Otto SJ, van der Kwast TH, de Koning HJ, Schröder FH ERSPC Rotterdam Study Group (2013) Screening for prostate cancer: results of the Rotterdam section of the european randomized study of screening for prostate cancer. Eu Urol 64: 530–539.
Taylor RA, Fraser M, Livingstone J, Espiritu SM, Thorne H, Huang V, Lo W, Shiah YJ, Yamaguchi TN, Sliwinski A, Horsburgh S, Meng A, Heisler LE, Yu N, Yousif F, Papargiris M, Lawrence MG, Timms L, Murphy DG, Frydenberg M, Hopkins JF, Bolton D, Clouston D, McPherson JD, van der Kwast T, Boutros PC, Risbridger GP, Bristow RG (2017) Germline BRCA2 mutations drive prostate cancers with distinct evolutionary trajectories. Nat Commun 9 (8): 13671.
Thompson D, Easton DF Breast Cancer Linkage Consortium (2002) Cancer Incidence in BRCA1 mutation carriers. J Natl Cancer Inst 94: 1358–1365.
Thorne H, Willems AJ, Niedermayr E, Hoh IM, Li J, Clouston D, Mitchell G, Fox S, Hopper JL Kathleen Cunningham Consortium for Research in Familial Breast Cancer Consortium, Bolton D (2011) Decreased prostate cancer-specific survival of men with BRCA2 mutations from multiple breast cancer families. Cancer Prev Res (Phila) 4: 1002–1010.
Tryggvadóttir L, Vidarsdóttir L, Thorgeirsson T, Jonasson JG, Olafsdóttir EJ, Olafsdóttir GH, Rafnar T, Thorlacius S, Jonsson E, Eyfjord JE, Tulinius H (2007) Prostate cancer progression and survival in BRCA2 mutation carriers. J Natl Cancer Inst 99: 929–935.
van Asperen CJ, Brohet RM, Meijers-Heijboer EJ, Hoogerbrugge N, Verhoef S, Vasen HF, Ausems MG, Menko FH, Gomez Garcia EB, Klijn JG, Hogervorst FB, van Houwelingen JC, van't Veer LJ, Rookus MA, van Leeuwen FE Netherlands Collaborative Group on Hereditary Breast Cancer (HEBON) (2005) Cancer risks in BRCA2 families: estimates for sites other than breast and ovary. J Med Genet 42: 711–719.
Vickers AJ, Till C, Tangen CM, Lilja H, Thompson IM (2011) An empirical evaluation of guidelines on prostate-specific antigen velocity in prostate cancer detection. J Natl Cancer Inst 103 (6): 462–469.
Acknowledgements
We are indebted to all of the men who are taking part in this study. This research is coordinated by the Institute of Cancer Research, London, UK and is supported by grants from Cancer Research UK (Grant references (C5047/A21332, C5047/A13232 and C5047/A17528) and The Ronald and Rita McAulay Foundation. Mr and Mrs Jack Baker for the study in NorthShore University HealthSystem, Evanston, Illinois and Myriad Genetics Laboratory, Salt Lake City, Utah, for providing research BRCA testing rates for NorthShore University HealthSystem participants. We acknowledge funding from the NIHR to the Biomedical Research Center at The Institute of Cancer Research and The Royal Marsden NHS Foundation Trust, at Central Manchester Foundation Trust and the Oxford Biomedical Research Centre Program. We acknowledge that in Australia, this project was co-funded by Cancer Council Tasmania and Cancer Australia, grant number 1006349 (2011–2013), Prostate Cancer Foundation of Australia, grant number PCFA PRO4 (2008) and Cancer Councils of Victoria and South Australia, grant number 400048 (2006–2008), The Victorian Cancer Agency Clinical Trial Capacity CTCB08_14, Cancer Australia & Prostate Cancer Foundation of Australia (2014–2016) grant number 1059423, and Translational grants EOI09_50. The Association of International Cancer Research funded data collection in The Netherlands (AICR 10–0596). We acknowledge funding from the Basser Center for BRCA (to S Domchek). We acknowledge funding from the National Cancer Institute [P30-CA008748], the Sidney Kimmel Center for Prostate and Urologic Cancers, and David H. Koch through the Prostate Cancer Foundation, the National Institute for Health Research (NIHR) Oxford Biomedical Research Centre Program in UK, Swedish Cancer Society (Cancerfonden project no. 11–0624), and the Swedish Research Council (VR-MH project no. 2016–02974). We acknowledge funding from the Slovenian Research Agency, Research programme P3–0352. Elena Castro acknolwedges funding from a Juan de la Cierva’ fellowship from MINIECO (grant reference IJCI- 2014–19129). We acknowledge the support of the Asociación Española Contra el Cáncer (AECC), the Instituto de Salud Carlos III (organismo adscrito al Ministerio de Economía y Competitividad) and ‘Fondo Europeo de Desarrollo Regional (FEDER), una manera de hacer Europa’ (PI10/01422, PI13/00285, PIE13/00022, PI16/00563 and CIBERONC) and the Institut Català de la Salut and Autonomous Government of Catalonia (2009SGR290, 2014SGR338 and PERIS Project MedPerCan). We acknowledge David Fisas, Consol Lopez and Dr Nuria Calvo for their involvement in the project at Hospital de Sant Pau, Barcelona. We are grateful to the members of the Data and Safety Monitoring Committee: S. Duffy (Chair), P. White (UK NEQAS representative) and J. McGrath (BAUS representative). We acknowledge the contribution of past members of the IMPACT Steering Committee: J. Melia, S. Moss, P. Wilson and G. Mitchell.
Author information
Authors and Affiliations
Consortia
Corresponding author
Ethics declarations
Competing interests
Hans Lilja holds patents for free PSA, hK2, and intact PSA assays, and is named, along with Andrew J. Vickers, on a patent application for a statistical method to detect prostate cancer. The marker assay patents and the patent application for the statistical model has been licensed and commercialised as the 4Kscore by OPKO Diagnostics. Drs Vickers and Lilja receive royalties from sales of this test. Additionally, Dr Lilja owns stock and Dr Vickers owns stock options in OPKO. Professor Rosalind Eeles—Janssen: provided medical education support to GU ASCO Feb 2013. Succinct Communications: received an honorarium and expenses for attending and speaking at UK Cancer Convention Oct 2013. The authors have no other conflict of interest to declare.
Appendix 1
Appendix 1
The IMPACT Collaborators
IMPACT Study Steering Committee
Rosalind Eeles—Institute of Cancer Research, London
Elizabeth Bancroft—Royal Marsden NHS Foundation Trust, London
Elizabeth Page—Institute of Cancer Research, London
Zsofia Kote-Jarai—Institute of Cancer Research, London
Audrey Ardern-Jones—Royal Marsden NHS Foundation Trust, London
Chris Bangma—Erasmus University Medical Center, Rotterdam
Elena Castro—CNIO, Madrid
David Dearnaley—Institute of Cancer Research, London
Alison Falconer—Imperial College Healthcare NHS Trust, London, UK
Christopher Foster—HCA Pathology Laboratories, London
Henrik Grönberg—University Hospital, Umea
Freddie C. Hamdy—University of Oxford, Oxford
Óskar Þór Jóhannsson—Landspitali—National University Hospital of Iceland, Reykjavik
Vincent Khoo—Royal Marsden NHS Foundation Trust, London
Diana Eccles—Wessex Clinical Genetics Service, Southampton
Hans Lilja—MSKCC, New York & University of Oxford, Oxford
Gareth Evans—St Mary’s Hospital, Manchester, UK
Jorunn Eyfjord—University of Iceland, Reykjavik
Jan Lubinski—International Hereditary Cancer Center, Szczecin
Lovise Maehle—Norwegian Radium Hospital, Oslo
Christos Mikropoulos—The Institute of Cancer Research
Alan Millner—Royal Marsden NHS Foundation Trust, London
Geoffrey Lindeman—Royal Melbourne Hospital; WEHI; Uni of Melbourne, Melbourne
Anita Mitra—University College London Hospitals, London
Sue Moss—Queen Mary University of London
Clare Moynihan—Institute of Cancer Research, London
Gad Rennert—CHS National Cancer Control Center, Carmel Medical Center, Haifa
Mohnish Suri—Nottingham City Hospital, Nottingham, UK
Penny Wilson—Director, BioZenix, Cheshire
Rosalind Eeles, Elizabeth Bancroft, Elizabeth Page, Sibel Saya, Alex Dias, Natalie Taylor, Kathryn Myhill, Sarah Thomas, Ashton Stroud, Jenny Pope, Anthony Chamberlain, Diana Keating—Coordinating Centre, Institute of Cancer Research, London
Australia (*more than 1 affiliation)
Gillian Mitchell*, Sue Shanley, Kate Richardson, Joanne McKinley, Lara Petelin, Morgan Murphy, Lyon Mascarenhas, Paul James*—Parkville Familial Cancer Centre, Peter MacCallum Cancer Centre, East Melbourne, VIC
Gillian Mitchell*, Paul James*—The Sir Peter MacCallum Department of Oncology, University of Melbourne, VIC
Paul James*—Genetics Medicine, Royal Melbourne Hospital, Melbourne, VIC
Declan Murphy—Department of Urology, Peter MacCallum Cancer Centre, East Melbourne, VIC
Jimmy Lam, Louise Taylor, Cathy Miller, Alan Stapleton, Michael Chong—Department of Urology, Repatriation General Hospital, Daw Park, SA
Graeme Suthers, Nicola Poplawski—SA Clinical Genetics Service, SA Pathology (at Women's & Children's Hospital), North Adelaide, SA
Katherine Tucker*, Lesley Andrews, Jessica Duffy—Hereditary Cancer Clinic, Prince of Wales Hospital, Randwick, NSW
Richard Millard—Department of Urology, Prince of Wales Hospital, Randwick, NSW
Robyn Ward, Rachel Williams—Hereditary Cancer Clinic, Prince of Wales Clinical School, Faculty of Medicine, UNSW, Sydney, NSW
Phillip Stricker—St Vincent's Clinic, Sydney, NSW
Judy Kirk*, Michelle Bowman—Familial Cancer Service, Westmead Hospital, Wentworthville, NSW
Judy Kirk*—Centre for Cancer Research, The Westmead Institute for Medical Research, NSW
Manish Patel—Westmead Hospital, Wentworthville, NSW
Marion Harris, Shona O'Connell, Clare Hunt, Courtney Smyth—Familial Cancer Centre, Monash Health, Clayton, VIC
Mark Frydenberg—Monash Medical Centre, VIC
Geoffrey Lindeman*, Kylie Shackleton, Catherine Morton—Parkville Familial Cancer Centre, The Royal Melbourne Hospital, Grattan St, Parkville, VIC
Geoffrey Lindeman*—Stem Cells and Cancer Division, The Walter and Eliza Hall Institute of Medical Research, Parkville, VIC
Geoffrey Lindeman*—Department of Medicine, The University of Melbourne, Parkville, VIC
Rachel Susman, Julie McGaughran, Melanie Boon—Genetic Health Queensland, Royal Brisbane & Women's Hospital, Herston, QLD
Nicholas Pachter*, Sharron Townshend, Lyn Schofield, Cassandra Nicholls—Genetic Services of WA, King Edward Memorial Hospital, Subiaco, WA
Nicholas Pachter*—Department of Paediatrics, University of Western Australia, Perth, WA
Allan Spigelman*, Margaret Gleeson—Hunter Family Cancer Service, Waratah, NSW
Allan Spigelman*—University of New South Wales, St Vincent’s Clinical School, NSW
Allan Spigelman*—Hereditary Cancer Clinic, The Kinghorn Cancer Centre, St Vincent's Hospital, Sydney, NSW
David Amor*, Jo Burke, Briony Patterson—Tasmanian Clinical Genetics Service, Hobart, TAS
David Amor*—Murdoch Childrens Research Institute, Parkville, VIC
Peter Swindle—Mater Private Hospitals, QLD
Rodney Scott—School of Biomedical Sciences and Pharmacy, University of Newcastle, NSW
Victorian Cancer Biobank, Carlton, VIC
Department Gynaecological Oncology Laboratory, Westmead Hospital, Centre for Cancer Research, NSW
PathWest (Clinical Trials Lab), Nedlands, WA
Pathology Queensland (Central Laboratory; Health Support Queensland), QLD Department of Health, Herston, QLD
Canada
William Foulkes, Talia Boshari, Armen Aprikian—McGill University, Montreal
Denmark
Thomas Jensen, Anders Bojeson, Palle Osther, Anne-Bine Skytte, Dorthe Cruger, Majbritt Kure Tøndering—Vejle Hospital, Vejle
Anne-Marie Gerdes—Odense University Hospital, Odense
Germany
Rita Schmutzler, Kerstin Rhiem, Petra Wihler—Center of Familial Breast and Ovarian Cancer, University Hospital of Cologne, Cologne
K Kast, C Griebsch—University Hospital Dresden
Iceland
Oskar Johannsson, Vigdis Stefansdottir—University Hospital of Iceland, Reykjavik
India
Vedang Murthy, Rajiv Sarin, Kasturi Awatagiri, Sujata Ghonge, Pradnya Kowtal, Gouri Mulgund—Tata Memorial Centre, Mumbai
Ireland
David Gallagher, Richard Bambury, Michael Farrell, Fergal Gallagher, Ingrid Kiernan—Mater Private Hospital, Dublin
Israel
Eitan Friedman—Chaim Sheba Medical Center, Tel-Hashomer and the Sackler School of Medicine, Tel-Aviv University, Tel-Aviv
Rakefet Chen-Shtoyerman, Alon Basevitch, Dan Leibovici, Ehud Melzer and Sagi Josefsberg Ben-Yehoshua—The Genetic Institute, the Gastroenterology Institute and the Urology Department, Kaplan Medical Centre, Rehovot
Italy
Nicola Nicolai, Paolo Radice, Riccardo Valdagni, Tiziana Magnani, Simona Gay—Istituto Nazionale dei Tumori, Milan
Malaysia
Soo Hwang Teo, Hui Meng Tan, Sook-Yee Yoon—Cancer Research Initiatives Foundation, Subang Jaya Medical Centre, Selangor Darul Ehsan
Soo Hwang Teo, Meow Keong Thong—University of Malaya, Kuala Lumpur
The Netherlands
STOET—Stichting Opsporing Erfelijke Tumoren, Leiden
Christi van Asperen—Leiden University Medical Centre, Leiden
Bart Kiemeney. Wendy van Zelst-Stams—Radboud University Nijmegen Medical Centre
Margreet G.E.M. Ausems, Rob B. Van der Luijt—University Medical Center Utrecht
Theo van Os—Academic Medical Center, Amsterdam
Mariëlle W.G. Ruijs—Netherlands Cancer Institute, Amsterdam
Muriel A. Adank—VU University Medical Center, Amsterdam
Rogier A. Oldenburg—Erasmus Medical Center, Rotterdam
A. (Paula) T.J.M. Helderman- van den Enden, B.A.H. Caanen—University Hospital Maastricht
Jan C. Oosterwijk—University Medical Centre Groningen
Norway
Lovise Maehle, Pal Moller, Bjorn Brennhovd, Heidi Medvik, Eldbjørg Hanslien—Norwegian Radium Hospital, Oslo
Poland
Cezary Cybulski, Jan Lubinski, Dominika Wokolorczyk—International Hereditary Cancer Centre, Szczecin
Portugal
Manuel Teixeira, Sofia Maia, Ana Peixoto, Rui Henrique, Jorge Oliveira, Nuno Gonçalves, Luís Araújo, Manuela Seixas, João Paulo Souto, Pedro Nogueira—Portuguese Oncology Institute, Porto
Slovakia
Lucia Copakova—National Cancer Institute, Bratislava
Slovenia
Janez Zgajnar, Mateja Krajc, Alenka Vrecar—Institute of Oncology, Ljubljana
Spain
Gabriel Capellá, Ignacio Blanco—Hereditary Cancer Program, Catalonian Institute of Oncology, Barcelona
Teresa Ramón y Cajal, David Fisas, Josefina Mora, Salvador Esquena—Hospital de Sant Pau, Barcelona
Judith Balmaña, Neus Gadea, Juan Morote—Hospital Vall d'Hebron, Barcelona
Sweden
Annelie Liljegren, Marie Hjälm -Eriksson, Karl-Johan Ekdahl, Stefan Carlsson—Karolinska University Hospital, Karolinska Institutet, Stockholm
United Kingdom
Angela George, Zoe Kemp, Jennifer Wiggins, Cathryn Moss, Vincent Khoo, Nicholas Van As, Alan Thompson, Chris Ogden, Christopher Woodhouse, Pardeep Kumar—Royal Marsden NHS Foundation Trust
D Gareth Evans, Barbara Bulman, Jeanette Rothwell, Karen Tricker—Manchester Regional Genetics Service, Manchester
Diana Eccles, Gillian Wise, Catherine Mercer, Donna McBride, Philandra Costello, Allison Pearce, Audrey Torokwa—Wessex Clinical Genetics Service, Southampton.
Marc Tischkowitz, Joan Paterson, Virginia Clowes, Amy Taylor, Barbara Newcombe—East Anglian Regional Genetics Service, Cambridge
Lisa Walker, Dorothy Halliday, Barbara Stayner, D Fleming-Brown—Oxford Regional Genetics Service, Oxford
Katie Snape, Helen Hanson, Shirley Hodgson, Glen Brice, Tessa Homfray, Carrie Hammond, Kelly Kohut, Uruj Anjum, Audrey Dearing, Mark Mencias—South West Thames Regional Genetics Service, London.
Carole Brewer, Alison Potter, Caroline Renton, Anne Searle, Kathryn Hill, Selina Goodman, Lynda Garcia, Gemma Devlin, Sarah Everest, Maria Nadolski—Peninsula Clinical Genetics Service. Exeter.
Alex Henderson, Fiona Douglas, Irene Jobson, Edgar Paez—Northern Clinical Genetics Service, Newcastle.
Alan Donaldson, Sue Tomkins—South West Regional Genetics Service, Bristol
Caroline Langman, Chris Jacobs, Gabriella Pichert, Adam Shaw, Anju Kulkarni, Vishakha Tripathi, Sarah Rose, Cecilia Compton, Michelle Watson and Cherylin Reinholtz—South East Thames Regional Genetics Service, Guys Hospital London
Angela Brady, Virginia Clowes, Huw Dorkins, Athalie Melville, Monika Kosicka-Slawinska, Carole Cummings, Vicki Kiesel, Marion Bartlett, Kashmir Randhawa, Natalie Ellery—North West Thames Regional Genetics Service, Harrow
Lucy Side, Alison Male, Kate Simon, Katie Rees, Cecilia Compton, Lizzie Tidey, Jana Gurasashvili—North East Thames Regional Genetics Service, NE Thames.
Jackie Cook, Louise Nevitt, Stuart Ingram, Alice Howell—North Trent Clinical Genetics Service, Sheffield
Derek Rosario, James Catto, Joanne Howson—Academic Urology Unit, Sheffield
Rachel Hart, Kai-Ren Ong, Cyril Chapman, Trevor Cole, Tricia Heaton, Jonathan Hoffman, Lucy Burgess, Wayne Glover, Camilla Huber—West Midlands Regional Clinical Genetics Service, Birmingham
Rosemarie Davidson, Mark Longmuir, Cathy Watt, Alexis Duncan—West of Scotland Genetics Service, Glasgow
Julian Barwell, Roger Kockelbergh, Shumikazi Mzazi, Amy Dineen, Ayisha Sattar, Beckie Kaemba, Zahirah Sidat, Nafisa Patel, Kas Siguake—Leicester Royal Infirmary
Alex Henderson, Angela Birt, Una Poultney, Nkem Umez-Eronini, Jaswant Mom—North Cumbria University Hospitals Trust
Lynn Greenhalgh, Vivienne Sutton—Cheshire and Mersey Clinical Genetics Service, Liverpool Women’s Hospital
Philip Cornford, Nicola Bermingham, Pembe Yesildag, Katy Treherne, Julie Griffiths—Royal Liverpool and Broadgreen Hospital NHS Trust, Liverpool
Carole Brewer, Lyn Cogley, Hannah Gott—Derriford Hospital, Plymouth
United States
Dr Wendy S Rubinstein, Dr Peter Hulick, Dr Michael McGuire, Dr Daniel Shevrin, Dr Karen Kaul, Scott Weissman CGC, Anna Newlin, Kristen Vogel, Shelly Weiss, Nicole Hook—NorthShore University HealthSystem, Evanston
Saundra Buys, David Goldgar, Tom Conner, Vickie Venne, Robert Stephenson, Christopher Dechet—Salt Lake City, Utah
Susan Domchek, Jacquelyn Powers, Neil Rustgi—University of Pennsylvania, Philadelphia
Sara Strom, Banu Arun, John W. Davis, Yuko Yamamura—MD Anderson, Texas
Elias Obeid, Veda Giri, Laura Gross, Linda Okoth—Fox Chase Cancer Center
Kathy Cooney, Elena Stoffel, Linda Oko—University of Michigan
Rights and permissions
This work is licensed under the Creative Commons Attribution-Non-Commercial-Share Alike 4.0 International License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Mikropoulos, C., Selkirk, C., Saya, S. et al. Prostate-specific antigen velocity in a prospective prostate cancer screening study of men with genetic predisposition. Br J Cancer 118, 266–276 (2018). https://doi.org/10.1038/bjc.2017.429
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2017.429
Keywords
This article is cited by
-
Screening for prostate cancer: protocol for updating multiple systematic reviews to inform a Canadian Task Force on Preventive Health Care guideline update
Systematic Reviews (2022)
-
The influence of BRCA2 mutation on localized prostate cancer
Nature Reviews Urology (2019)
-
Cancer Genomics for Oncologists: Cancer Risk and Management of BRCA1 and BRCA2 Carriers
Current Genetic Medicine Reports (2019)
-
Risk and Prevention for Highly Penetrant Genes
Current Breast Cancer Reports (2018)
-
Aussagekraft des PSA bei BRCA-Mutation
InFo Onkologie (2018)