Abstract
Background:
Altered cellular metabolism is a hallmark of cancer and some are reliant on glutamine for sustained proliferation and survival. We hypothesise that the glutamine–proline regulatory axis has a key role in breast cancer (BC) in the highly proliferative classes.
Methods:
Glutaminase (GLS), pyrroline-5-carboxylate synthetase (ALDH18A1), and pyrroline-5-carboxylate reductase 1 (PYCR1) were assessed at DNA/mRNA/protein levels in large, well-characterised cohorts.
Results:
Gain of PYCR1 copy number and high PYCR1 mRNA was associated with Luminal B tumours. High ALDH18A1 and high GLS protein expression was observed in the oestrogen receptor (ER)+/human epidermal growth factor receptor (HER2)– high proliferation class (Luminal B) compared with ER+/HER2– low proliferation class (Luminal A) (P=0.030 and P=0.022 respectively), however this was not observed with mRNA. Cluster analysis of the glutamine–proline regulatory axis genes revealed significant associations with molecular subtypes of BC and patient outcome independent of standard clinicopathological parameters (P=0.012). High protein expression of the glutamine–proline enzymes were all associated with high MYC protein in Luminal B tumours only (P<0.001).
Conclusions:
We provide comprehensive clinical data indicating that the glutamine–proline regulatory axis plays an important role in the aggressive subclass of luminal BC and is therefore a potential therapeutic target.
Similar content being viewed by others
Main
Deregulation of metabolic pathways has been readily accepted as part of the revised hallmarks of cancer, where cancer cells are able to regulate their metabolism to provide energy and cellular building blocks required for growth (Hanahan and Weinberg, 2011). Many cancer cells are highly reliant on amino acids for their growth where endogenous synthesis may not provide the rapidly proliferating cells with sufficient nutrients for nuclear biosynthesis. There is also increasing evidence that oncogenes and/or tumour-suppressor genes can reprogram tumour cell metabolism including the direct regulation of the glutamine (Gln)–proline (Pro) regulatory axis by MYC and p53 (Green et al, 2016; Hu et al, 2010; Polyak et al, 1997). This axis is the most important metabolic pathway in tumours after glucose primarily as Gln is used to replenish the tricarboxylic acid (TCA) cycle and supplies carbon and nitrogen for synthesis of nucleotides, amino acids and glutathione. Indeed, some solid tumours have Gln-dependent cell growth or ‘glutamine addiction’ (Wise and Thompson, 2010).
Glutamine is a non-essential amino acid synthesised by glutamine synthetase (GS) from glutamate and ammonia. Its utilisation, via reductive carboxylation, is necessary for sustained proliferation/survival and is linked with resistance to certain drugs (Alberghina & Gaglio, 2014). A further role for Gln in cancer cell protein translation stems from observations that a master regulator of protein translation, rapamycin complex 1 (mTORC1), which regulates cell growth and protein translation, is also responsive to Gln levels (Csibi et al, 2013). In breast cancer (BC), high-grade highly proliferative tumours such as the triple negative (TN), have higher levels of glutamate and glutaminase (GLS) together with low levels of Gln than low-grade tumours and normal breast epithelium (Budczies et al, 2015; Gross et al, 2014; Kim et al, 2013). Metabolic profiles of BC show glutaminolysis metabolism as a key pathway discriminating between TN and oestrogen receptor (ER)+ tumours (Cao et al, 2014). The CB-839 small-molecule selective inhibitor of GLS, which has anti-tumour activity in TNBC cell lines is currently being tested in phase I clinical trials (NCT02071862) (Gross et al, 2014).
Glutamine is converted to glutamate by GLS before entering the TCA cycle as a precursor to α-ketoglutarate, an important energy source which is synthesised by glutamate dehydrogenase. However, glutamate can also be converted into Pro via the enzymes pyrroline-5-carboxylate synthetase (ALDH18A1) and pyrroline-5-carboxylate reductase 1 (PYCR1), which subsequently produces NADP used to fuel the breakdown of glucose by glycolysis (Liu & Phang, 2012; Phang et al, 2015) (Figure 1). ALDH18A1 is responsible for reducing glutamate to Gln semialdehyde; a crucial step in the de novo biosynthesis of Pro.
With renewed interest in oncometabolism, metabolic enzymes are increasingly targeted to improve therapeutic efficacy and reduce resistance. We therefore hypothesised that the Pro–Gln axis is a key metabolic pathway regulated by MYC in BC particularly as we have recently showed Pro dehydrogenase (PRODH) to be down regulated (Green et al, 2016). This pathway could be used as a potential therapeutic target particularly as the pleotropic MYC has so far proved ineffective.
The aim of this study was to conduct a comprehensive genomic, transcriptomic and proteomic analysis of the Gln–Pro regulatory axis in the molecular subtypes of BC and their associations with MYC. More specifically, we aim to determine GLS, ALDH18A1 and PYCR1 gene copy number and gene expression; together with its protein expression, in large well-characterised annotated cohorts of BC to determine their biological, clinicopathological and prognostic value in the different molecular classes with particular interest in the highly proliferative aggressive subgroups as potential therapeutic targets.
Materials and methods
Gln–Pro enzymes copy number and gene expression
PYCR1, ALDH18A1, GLS and MYC copy number and gene expression were evaluated in a cohort of 1980 BC samples using the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) cohort (Curtis et al, 2012). METABRIC provides data on genomic and transcriptomic profiling of BC using the Affymetrix SNP 6.0 and Illumina HT-12 v3 platforms, respectively. In addition, TP53 mutational profiling was performed. Detailed description of the experimental assays and analytical methods used were described previously. In this cohort, patients with ER+ and/or lymph node negative tumours did not receive adjuvant chemotherapy, while those with ER− and/or lymph node positive tumours received adjuvant chemotherapy. Breast cancer-specific survival is defined as the time (in months) from the date of primary surgery to the date of BC-related death.
Gln–Pro enzymes protein expression
Immunohistochemistry was conducted for PYCR1, ALDH18A1 and GLS using a large cohort of patients comprising a well-characterised consecutive series of early stage (TNM stage I–III excluding T3 and T4 tumours) sporadic primary operable invasive BC. Patients (age ⩽70 years) were enroled into the Nottingham Tenovus Primary Breast Carcinoma Series, presented at Nottingham City Hospital between 1989 and 1998 (n=1837) and managed in accordance to uniform protocol. Patients’ clinical history, tumour characteristics, information on therapy and outcomes are prospectively maintained. Outcome data were collected on a prospective basis.
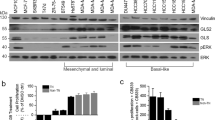
Primary antibody specificity (ALDH18A1 1 : 250 (HPA012604, Sigma-Aldrich, UK), GLS 1 : 1000 (EP7212, Abcam, UK) PYCR1 1 : 250 (HPA047660, Sigma-Aldrich, UK) was validated by western blotting using MCF7, MDA-MB-231 and SKBR3 human BC cell lines (American Type Culture Collection; Rockville, MD, USA). Proteins were detected using IRDye 800CW and 680RD fluorescent secondary antibodies (1 : 15 000 dilution. 926-32213 and 926-68072, LI-COR Biosciences) and visualised using the Odyssey Fc with Image Studio 4.0 (LI-COR Biosciences). Anti-β-actin primary antibody (Sigma-Aldrich) was used as a loading control (1 : 5000). Specific bands were observed at the correct molecular weights for ALDH18A1 (87 kDa), GLS (73, 65 kDa) and PYCR1 (36 kDa). An additional 55 kDa band was observed for GLS and a 28 kDa band for PYCR1, which represent alternate isoforms of the proteins.
Tissue arrays and immunohistochemistry
Tumour samples, 0.6 mm cores, were arrayed as previously described (Abd El-Rehim et al, 2005). Immunohistochemical staining was performed on 4 μm thick sections using the Novolink polymer detection system (Leica Biosystems, RE7150-K), and following the manufacturer’s protocol. Primary antibodies for PYCR1, ALDH18A1 and GLS were diluted at 1 : 50 in Leica antibody diluent (RE7133). Negative (omission of primary antibody) and positive controls were included according to manufacturer’s data sheet.
Stained TMA sections were scored using high-resolution digital images (NanoZoomer; Hamamatsu Photonics, Welwyn Garden City, UK) at × 20 magnification. Staining was assessed using the semi-quantitative, modified histochemical score (H-score), which evaluates both the intensity of staining and the percentage of stained cells resulting in a final score of 0–300 (Dang, 2012). For intensity, a score index of 0, 1, 2, and 3 corresponding to negative, weak, moderate, and strong was used and the percentage of positive cells for each intensity was estimated subjectively. Dichotomisation of PYCR1, ALDH18A1 and GLS protein expression was determined using the median H-score. All cores were scored by NJ or HC and a pathologist (MA), blinded from the scores and the clinical data, scored 10% of cores for inter-observer concordance. There was high inter and intra-observer concordance between the scorers (Kappa score ⩾0.6).
Immunohistochemical staining and dichotomisation of other biomarkers included in this study were as per previous publications (Green et al, 2016). Oestrogen receptor (ER) and PgR positivity was defined as ⩾1% staining. Immunoreactivity of human epidermal growth factor receptor (HER2) in TMA cores was scored using standard HercepTest guidelines (Dako). Chromogenic in situ hybridisation (CISH) was used to quantify HER2 gene amplification in borderline cases using the HER2 FISH pharmDx plus HER2 CISH pharmDx kit (Dako) and was assessed according to the American Society of Clinical Oncology guidelines. Breast cancer molecular subtypes were defined based on the IHC profile as: Luminal A: ER+/HER2− low proliferation (Ki67<10%), Luminal B: ER+/HER2− high proliferation (Ki67⩾10%), HER2-positive class: HER2+ regardless of ER status, TN: ER−, PgR− and HER2−. Basal phenotype was defined as those tumours expressing cytokeratin (Ck) 5/6, and/or Ck14 and/or Ck17.
Cluster analysis
The partitioning around medoids (PAM) algorithm (also known as k-medoids algorithm) was used to cluster tumours based on gene and protein expression of the Gln–Pro enzymes as previously described (Soria et al, 2010). A number of cluster validity indices were used to determine the best number of clusters as the explicit input parameter to the PAM algorithm (Soria et al, 2010).
Statistical analysis
Statistical analysis was performed using SPSS 21.0 statistical software (SPSS Inc., Chicago, IL, USA). Univariate and multivariate analyses were performed by χ2-test, Log rank and Cox regression analysis, respectively. One way ANOVA (Tukey) and Spearman’s correlation coefficient were used for continuous data. Survival curves were analysed by Kaplan–Meier. A P-value <0.05 was considered significant. This study complied with reporting recommendations for tumour marker prognostic studies (REMARK) criteria (McShane et al, 2005).
Ethics
This study was approved by the Nottingham Research Ethics Committee 2 under the title ‘Development of a molecular genetic classification of breast cancer’. All samples from Nottingham used in this study were pseudo-anonymised and collected prior to 2006 and therefore under the Human Tissue Act informed patient consent was not needed. Release of data was also pseudo-anonymised as per Human Tissue Act regulations.
Results
Gln–Pro regulatory axis expression in breast cancer
Gain of PYCR1 copy number primarily occurred in Luminal B tumours with 116 out of 257 (45%) of all gains occurring (Table 1, P<0.001). Expression of PYCR1 mRNA was significantly higher in Luminal B tumours compared with Luminal A tumours, and high expression was also observed with HER2+ and Basal/TN subtypes (Figure 2A, P<0.001). However there was no association observed between PYCR1 protein expression and molecular classes (Figure 2B, P=0.118).
Within the molecular classes, high ALDH18A1 mRNA was associated with HER2+ tumours although there was no difference observed with mRNA levels between Luminal A and Luminal B tumours (Figure 2C, P<0.001). Copy number gain of ALDH18A1 was associated with basal tumours (Table 1, P<0.001). In contrast, high ALDH18A1 protein expression was seen in the ER+/HER2− high proliferation class compared with ER+/HER2− low proliferation tumours (P=0.030); however significance was not observed with all molecular classes (Figure 2D; P=0.079).
High copy number gain of GLS (Table 1), GLS mRNA (Figure 2E) and protein (Figure 2F) were all significantly associated with basal/TN subtypes (P<0.001). Lower GLS mRNA expression was seen in Luminal B tumours compared with Luminal A and the other molecular subtypes (Figure 2E; P<0.001). In contrast, higher protein expression of GLS was seen in ER+/HER2− high proliferation compared with those classified as ER+/HER2− low proliferation (Figure 2F; P=0.022), where highest expression was seen in the TN tumours (P<0.001).
Gln–Pro regulatory axis confers poor prognosis in Luminal B breast cancer
Breast tumours were further clustered based on the comparison of several indices comparing the gene expression of the Gln–Pro enzymes. These were characterised as follows: Cluster 1 (GLS−/ALDH18A1+/PYCR1+), Cluster 2 (GLS+/ALDH18A1+/PYCR1+) and Cluster 3 (GLS+/ALDH18A1−/PYCR1−) (Figure 3A). The clusters were significantly associated with molecular subtypes of BC, where Cluster 1 were predominately Luminal B and HER2+ tumours, Cluster 2 were associated with basal/HER2+ tumours and Cluster 3 were primarily Luminal A and normal subtypes (Table 2). With respect to patient outcome, Cluster 1 tumours had the worst survival compared with Cluster 2, which had moderate outcome and Cluster 3 which showed the best survival (Figure 3B; P<0.001). In Cox regression, Gln–Pro PAM clusters remained independent of the standard clinicopathological parameters in predicting patient survival (Table 3; P=0.012).
We looked to replicate the clusters derived from the mRNA expression using the protein expression of the Gln–Pro enzymes (using the median H-scores), which similarly showed that Cluster 1 (GLS−/ALDH18A1+/PYCR1+) was strongly associated with ER+/HER2− high proliferation tumours, whereas Cluster 2 (GLS+/ALDH18A1+/PYCR1+) and Cluster 3 (GLS+/ALDH18A1−/PYCR1−) were associated with ER+/HER2− low proliferation and TN/HER2+ tumours, respectively (Table 2; P=0.001). In terms of patient outcome, Cluster 1 tumours showed the worst survival compared with Clusters 2 and 3 (Figure 3C; P=0.0003). In Cox regression analysis, the Glu–Pro Clusters remained independent of tumour grade, lymph node stage, tumour size, ER and HER2 status (Table 3; P=0.001).
c-MYC is associated with high Gln–Pro enzymes in Luminal B tumours
MYC mRNA was negatively correlated with PCYR1 (P=0.01) and ALDH18A1 (P<0.001) in all breast tumours, but not GLS (Table 4). In specific subtypes, MYC was positively correlated with PYCR1 mRNA in Luminal B tumours (P=0.006) and negatively correlated in Luminal A tumours (Table 4; P<0.001). The only other correlations between MYC and the Gln–Pro genes was a negative correlation with ALDH18A1 (P<0.001) in Luminal A (P<0.001) and HER2+ tumours (P=0.002).
In terms of protein expression, MYC protein was positively associated with the individual protein expression of GLS and ALDH18A1 in all BCs, but was only associated with all three Gln–Pro enzymes in ER+/HER2− high proliferation tumours (P<0.001, Table 4).
Discussion
ER+/luminal tumours, which comprise the majority of BC (55–80%), remain a heterogeneous group in terms of molecular biology and patients’ outcome. Despite the significant benefit of hormone therapy, a proportion of patients with luminal BC develop recurrences and die of their disease. There is therefore a clear need for improved understanding of the biology of the luminal class of BC, with subsequent translation into more effective methods of prognostic and predictive stratification of this most common form of BC.
Metabolic reprogramming in cancer including BC provides a vital role in the provision of supplementary elements including nutrients and energy, which are essential for cellular growth. Tumour cells can become reliant on Gln metabolism and become ‘addicted’ to this amino acid for sustained proliferation/survival. Additionally, Pro is often used as a precursor for other amino acids; therefore it is important in the synthesis of new proteins, as well as mediating redox signalling and protecting cells from oxidative stress (Phang et al, 2015; Kuo et al, 2016).
It has also been shown that there is a major metabolic shift towards de novo Pro synthesis in metastatic breast tumours (Richardson et al, 2008). High levels of MYC are required to maintain this glutaminolytic phenotype, which not only sees an increase in Gln cellular uptake but also regulates Gln–Pro enzymes (Wise et al, 2008; Gao et al, 2009). MYC promotes the conversion of Gln to Pro by nearly 10-fold by upregulating ALDH18A1 (Hu et al, 2008; Liu et al, 2012).
Evidence of Gln dependence in TNBC has previously been established (Hanahan and Weinberg, 2011; Cao et al, 2014; Gross et al, 2014; Lukey et al, 2016) but studies that address the prognostic significance of the key Gln–Pro enzymes in BC and their potential influence on Gln metabolism in the other molecular subtypes remains limited particularly in luminal BC. We have therefore investigated the expression of PYCR1, ALDH18A1 and GLS at the genomic, transcriptomic and proteomic level, and the impact of MYC on their expression, utilising a large number of breast tumours in order to better understand the potential role of this regulatory axis in BC and its molecular subtypes, particularly in the luminal tumours.
Overexpression of ALDH18A1 increases Pro levels and lowers reactive oxygen species, as well as increasing cell survival. The increase in Pro has been linked to protect cells against hydrogen peroxide-induced cell death, as well as carcinogenic stressors; therefore promoting tumour growth and proliferation (Krishnan et al, 2008). In line with this, we show that high ALDH18A1 confers a poor prognosis in Luminal B tumours. Inhibition of ALDH18A1 in melanoma significantly disrupts Pro synthesis limiting cellular metabolism and decreasing cell viability, tumour growth and protein synthesis (Liu et al, 2012; Kardos et al, 2015). Knockdown or inhibition of ALDH18A1 in Luminal B tumours could therefore potentially lead to a benefit in therapeutic targeting of this enzyme.
PYCR1 is frequently overexpressed in many cancer types (Kuo et al, 2016); in particular breast, pancreatic and ovarian cancer (Natarajan et al, 2012; Phang et al, 2015). In meta-analysis, PYCR1 mRNA is highly expressed in the more aggressive BC subtypes, in addition to its protein in a small number of BC cases (Ding et al, 2017). While we show similar results for mRNA in the METABRIC data set, we were unable to confirm the translation into similarly high protein expression in TNBC and HER2+ BC nor its association with patient outcome. However, we do show that Luminal B have a high copy number gain of PYCR1 and consequently supports a role in this poor prognostic group of tumours. In prostate cancer, knockdown of PYCR1 results in the inhibition of cell proliferation via cell cycle arrest and enhanced apoptosis (Zeng et al, 2017) although the effect on BC cells, particularly Luminal B tumours remains to be determined.
Levels and activity of GLS are significantly increased in TNBC and HER2+ BC cell lines due to their propensity for Gln dependence (Wang et al, 2010; Gross et al, 2014; Lukey et al, 2016) which we here confirm in vivo. However, we further demonstrate that GLS predicts a better outcome in HER2+ tumours and selective GLS inhibitors, such as CB-839 which blocks Gln consumption and reduces subsequent glutamate-derived metabolic intermediates, is likely to have limited use in these patients (Gross et al, 2014).
Protein analysis also revealed that PYCR1 and ALDH18A1 are significantly associated with the MYC oncogene. This coincides with a plethora of studies, which have shown that the MYC oncogene reprograms cell metabolism and causes an upregulation of genes involved in cancer (Liu et al, 2012; Li and Simon, 2013; Phang et al, 2013). Multiple studies have also supported the findings in this study, as the MYC oncogene has resulted in an increased expression of PYCR1 and ALDH18A1 to drive Pro production; alongside fuelling the Gln and MYC addiction that cancer cells exhibit (Wise et al, 2008; Li and Simon, 2013; Phang et al, 2015).
It is thought that the MYC gene interacts with these enzymes by amplifying the existing transcriptional signalling of the gene leading to an increase in mRNA. MYC then drives the translation of mRNA in order to increase protein levels of the enzymes (Natarajan et al, 2012; Li and Simon, 2013). Interestingly, studies by Wei Liu et al have shown that downregulating the MYC oncogene with siRNAs not only reduced the conversion of Gln to Pro but also resulted in reduced mRNA and protein expression of ALDH18A1 and PYCR1 (Liu et al 2012, 2015). Silencing MYC results in a reduction of cell proliferation and induction of apoptosis; as well as decreasing levels of PYCR1 and ALDH18A1 directly (Dang, 2012; Liu et al, 2012) and targeting all three enzymes led to the substantial reduction in Pro production (Liu et al, 2012).
Our observational findings of the Gln–Pro enzymes and their differing associations with MYC within the subgroups of BC further support the many functions of MYC within glutaminolysis. We have previously shown that MYC is potentially driving glucose metabolism in ER-negative tumours and translational function in ER-positive tumours (Green et al, 2016), and we further show in this study that MYC is likely driving the conversion of Gln to Pro within Luminal B tumours. Pro metabolism has a functional role in redox regulation and the conversion of Gln to Pro could be one mechanism, where this balance can be achieved, especially in the highly proliferative Luminal B tumours, which show increased metabolic rates resulting in the accumulation of ROS (Benassi et al, 2006).
In this study, we have shown that the Gln–Pro enzymes are highly expressed in a subset of ER+ tumours that have high proliferation, that is, Luminal B tumours, and are related with poor patient outcome in this group, suggesting that expression of these enzymes are regulated and driven by alternative mechanisms within the different molecular subtypes of BC and hence the pathways utilised for cell metabolism will also differ. It was also interesting to observe the difference in gene and protein expression of these enzymes within the ER+/HER2− low proliferation tumours and TNBC/HER2+ tumours. This could have arisen due to the definitions used between mRNA (PAM50) and protein (ER plus Ki67 expression). It might also reflect the differences in transcription and translational modifications.
The high expression of the Gln–Pro regulatory axis in Luminal B tumours is perhaps not unsurprising, as they will have heavier demands of nutrients and energy essential for cell survival and proliferation compared with Luminal A tumours and it is known that tumours will alter their metabolic profiles to meet the needs for their growth and proliferation. This is supported by a study carried out by Kim et al (2013) in which differential expression patterns of Gln metabolism-related proteins were identified according to the molecular subtype of BC, with HER2+ tumours displaying highest Gln metabolic activity and higher MYC amplification and Luminal B tumours displaying higher Gln metabolic activity than Luminal A tumours.
We further show that within the highly proliferative Luminal B tumours, high expression of the Gln–Pro enzymes are all significantly associated with MYC, suggesting that it is the driving force behind the metabolic status for this subclass of BC.
Therefore, we believe that continued refinement in understanding of the biological diversity of BC, particularly the Luminal B class, with linked development of classification strategies suitable for routine clinical use are essential to achieve a personalised approach to BC management. Further investigation of Gln metabolism using a variety of in vitro functional assays are therefore essential to assess its the potential therapeutic value in the highly proliferative aggressive subclass of ER+Luminal B BC.
Change history
23 January 2018
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Abd El-Rehim DM, Ball G, Pinder SE, Rakha E, Paish C, Robertson JF, Macmillan D, Blamey RW, Ellis IO (2005) High-throughput protein expression analysis using tissue microarray technology of a large well-characterised series identifies biologically distinct classes of breast cancer confirming recent cDNA expression analyses. Int J Cancer 116 (3): 340–3450.
Alberghina L, Gaglio D (2014) Redox control of glutamine utilization in cancer. Cell death & disease 5: e1561.
Benassi B, Fanciulli M, Fiorentino F, Porrello A, Chiorino G, Loda M, Zupi G, Biroccio A (2006) c-Myc phosphorylation is required for cellular response to oxidative stress. Mol Cell 21 (4): 509–519.
Budczies J, Pfitzner BM, Gyorffy B, Winzer KJ, Radke C, Dietel M, Fiehn O, Denkert C (2015) Glutamate enrichment as new diagnostic opportunity in breast cancer. Int J Cancer 136 (7): 1619–1628.
Cao MD, Lamichhane S, Lundgren S, Bofin A, Fjosne H, Giskeodegard GF, Bathen TF (2014) Metabolic characterization of triple negative breast cancer. BMC Cancer 14: 941.
Csibi A, Fendt SM, Li C, Poulogiannis G, Choo AY, Chapski DJ, Jeong SM, Dempsey JM, Parkhitko A, Morrison T, Henske EP, Haigis MC, Cantley LC, Stephanopoulos G, Yu J, Blenis J (2013) The mTORC1 pathway stimulates glutamine metabolism and cell proliferation by repressing SIRT4. Cell 153 (4): 840–854.
Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, Graf S, Ha G, Haffari G, Bashashati A, Russell R, McKinney S, Group M, Langerod A, Green A, Provenzano E, Wishart G, Pinder S, Watson P, Markowetz F, Murphy L, Ellis I, Purushotham A, Borresen-Dale AL, Brenton JD, Tavare S, Caldas C, Aparicio S (2012) The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486 (7403): 346–352.
Dang CV (2012) MYC on the path to cancer. Cell 149 (1): 22–35.
Ding J, Kuo ML, Su L, Xue L, Luh F, Zhang H, Wang J, Lin TG, Zhang K, Chu P, Zheng S, Liu X, Yen Y (2017) Human mitochondrial pyrroline-5-carboxylate reductase 1 promotes invasiveness and impacts survival in breast cancers. Carcinogenesis 38: 519–531.
Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, Zeller KI, De Marzo AM, Van Eyk JE, Mendell JT, Dang CV (2009) c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature 458 (7239): 762–765.
Green AR, Aleskandarany MA, Agarwal D, Elsheikh S, Nolan CC, Diez-Rodriguez M, Macmillan RD, Ball GR, Caldas C, Madhusudan S, Ellis IO, Rakha EA (2016) MYC functions are specific in biological subtypes of breast cancer and confers resistance to endocrine therapy in luminal tumours. Br J Cancer 114 (8): 917–928.
Gross MI, Demo SD, Dennison JB, Chen L, Chernov-Rogan T, Goyal B, Janes JR, Laidig GJ, Lewis ER, Li J, Mackinnon AL, Parlati F, Rodriguez ML, Shwonek PJ, Sjogren EB, Stanton TF, Wang T, Yang J, Zhao F, Bennett MK (2014) Antitumor activity of the glutaminase inhibitor CB-839 in triple-negative breast cancer. Mol Cancer Ther 13 (4): 890–901.
Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144 (5): 646–674.
Hu CA, Khalil S, Zhaorigetu S, Liu Z, Tyler M, Wan G, Valle D (2008) Human Delta1-pyrroline-5-carboxylate synthase: function and regulation. Amino Acids 35 (4): 665–672.
Hu W, Zhang C, Wu R, Sun Y, Levine A, Feng Z (2010) Glutaminase 2, a novel p53 target gene regulating energy metabolism and antioxidant function. Proc Natl Acad Sci USA 107 (16): 7455–7460.
Kardos GR, Wastyk HC, Robertson GP (2015) Disruption of proline synthesis in melanoma inhibits protein production mediated by the GCN2 pathway. Mol Cancer Res 13 (10): 1408–1420.
Kim S, Kim DH, Jung WH, Koo JS (2013) Expression of glutamine metabolism-related proteins according to molecular subtype of breast cancer. Endocr Relat Cancer 20 (3): 339–348.
Krishnan N, Dickman MB, Becker DF (2008) Proline modulates the intracellular redox environment and protects mammalian cells against oxidative stress. Free Radic Biol Med 44 (4): 671–681.
Kuo ML, Lee MB, Tang M, den Besten W, Hu S, Sweredoski MJ, Hess S, Chou CM, Changou CA, Su M, Jia W, Su L, Yen Y (2016) PYCR1 and PYCR2 interact and collaborate with RRM2B to protect cells from overt oxidative stress. Sci Rep 6: 18846.
Li B, Simon MC (2013) Molecular pathways: targeting MYC-induced metabolic reprogramming and oncogenic stress in cancer. Clin Cancer Res 19 (21): 5835–5841.
Liu W, Hancock CN, Fischer JW, Harman M, Phang JM (2015) Proline biosynthesis augments tumor cell growth and aerobic glycolysis: involvement of pyridine nucleotides. Sci Rep 5: 17206.
Liu W, Le A, Hancock C, Lane AN, Dang CV, Fan TW, Phang JM (2012) Reprogramming of proline and glutamine metabolism contributes to the proliferative and metabolic responses regulated by oncogenic transcription factor c-MYC. Proc Natl Acad Sci USA 109 (23): 8983–8988.
Liu W, Phang JM (2012) Proline dehydrogenase (oxidase) in cancer. BioFactors 38 (6): 398–406.
Lukey MJ, Greene KS, Erickson JW, Wilson KF, Cerione RA (2016) The oncogenic transcription factor c-Jun regulates glutaminase expression and sensitizes cells to glutaminase-targeted therapy. Nat Commun 7: 11321.
McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM Statistics Subcommittee of the NCIEWGoCD (2005) REporting recommendations for tumour MARKer prognostic studies (REMARK). Br J Cancer 93 (4): 387–391.
Natarajan SK, Zhu W, Liang X, Zhang L, Demers AJ, Zimmerman MC, Simpson MA, Becker DF (2012) Proline dehydrogenase is essential for proline protection against hydrogen peroxide induced cell death. Free Radic Biol Med 53 (5): 1181–1191.
Phang JM, Liu W, Hancock C (2013) Bridging epigenetics and metabolism: role of non-essential amino acids. Epigenetics 8 (3): 231–236.
Phang JM, Liu W, Hancock CN, Fischer JW (2015) Proline metabolism and cancer: emerging links to glutamine and collagen. Curr Opin Clin Nutr Metab Care 18 (1): 71–77.
Polyak K, Xia Y, Zweier JL, Kinzler KW, Vogelstein B (1997) A model for p53-induced apoptosis. Nature 389 (6648): 300–305.
Richardson AD, Yang C, Osterman A, Smith JW (2008) Central carbon metabolism in the progression of mammary carcinoma. Breast Cancer Res Treat 110 (2): 297–307.
Soria D, Garibaldi JM, Ambrogi F, Green AR, Powe D, Rakha E, Macmillan RD, Blamey RW, Ball G, Lisboa PJ, Etchells TA, Boracchi P, Biganzoli E, Ellis IO (2010) A methodology to identify consensus classes from clustering algorithms applied to immunohistochemical data from breast cancer patients. Comput Biol Med 40 (3): 318–330.
Wang JB, Erickson JW, Fuji R, Ramachandran S, Gao P, Dinavahi R, Wilson KF, Ambrosio AL, Dias SM, Dang CV, Cerione RA (2010) Targeting mitochondrial glutaminase activity inhibits oncogenic transformation. Cancer cell 18 (3): 207–219.
Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, Nissim I, Daikhin E, Yudkoff M, McMahon SB, Thompson CB (2008) Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci USA 105 (48): 18782–18787.
Wise DR, Thompson CB (2010) Glutamine addiction: a new therapeutic target in cancer. Trends Biochem Sci 35 (8): 427–433.
Zeng T, Zhu L, Liao M, Zhuo W, Yang S, Wu W, Wang D (2017) Knockdown of PYCR1 inhibits cell proliferation and colony formation via cell cycle arrest and apoptosis in prostate cancer. Med Oncol 34 (2): 27.
Acknowledgements
We thank the Nottingham Health Science Biobank and Breast Cancer Now Tissue Bank for the provision of tissue samples. We thank the University of Nottingham (Nottingham Life Cycle 6 and Cancer Research Priority Area) for funding.
Author contribution
ARG & MLC conceived and designed study. HC, NJ, NDMC, KWC, MAA, RE-A, MD-R, CCN, IOE, EAR, ARG carried out experiments and collected data. MLC, HC, NJ, KWC, DS, ARG analysed data. All authors were involved in writing the paper and had final approval of the submitted and published versions.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Craze, M., Cheung, H., Jewa, N. et al. MYC regulation of glutamine–proline regulatory axis is key in luminal B breast cancer. Br J Cancer 118, 258–265 (2018). https://doi.org/10.1038/bjc.2017.387
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2017.387
Keywords
This article is cited by
-
Targeting glutamine metabolism improves sarcoma response to radiation therapy in vivo
Communications Biology (2024)
-
HuR controls glutaminase RNA metabolism
Nature Communications (2024)
-
Exploration of subtype-specific perturbations in breast cancer
Journal of Proteins and Proteomics (2024)
-
Polyamines: the pivotal amines in influencing the tumor microenvironment
Discover Oncology (2024)
-
Glutaminase (GLS1) gene expression in primary breast cancer
Breast Cancer (2023)