Abstract
Background:
Histopathological prognostication relies on morphological pattern recognition, but as numbers of biomarkers increase, human prognostic pattern-recognition ability decreases. Follicular lymphoma (FL) has a variable outcome, partly determined by FOXP3 Tregs. We have developed an automated method, hypothesised interaction distribution (HID) analysis, to analyse spatial patterns of multiple biomarkers which we have applied to tumour-infiltrating lymphocytes in FL.
Methods:
A tissue microarray of 40 patient samples was used in triplex immunohistochemistry for FOXP3, CD3 and CD69, and multispectral imaging used to determine the numbers and locations of CD3+, FOXP3/CD3+ and CD69/CD3+ T cells. HID analysis was used to identify associations between cellular pattern and outcome.
Results:
Higher numbers of CD3+ (P=0.0001), FOXP3/CD3+ (P=0.0031) and CD69/CD3+ (P=0.0006) cells were favourable. Cross-validated HID analysis of cell pattern identified patient subgroups with statistically significantly different survival (35.5 vs 142 months, P=0.00255), a more diffuse pattern associated with favourable outcome and an aggregated pattern with unfavourable outcome.
Conclusions:
A diffuse pattern of FOXP3 and CD69 positivity was favourable, demonstrating ability of HID analysis to automatically identify prognostic cellular patterns. It is applicable to large numbers of biomarkers, representing an unsupervised, automated method for identification of undiscovered prognostic cellular patterns in cancer tissue samples.
Similar content being viewed by others
Main
Follicular lymphoma (FL) follows an indolent clinical course in the majority of patients although outcome is highly variable (Freedman, 2012). Tumour histological grade and FLIPI score (Solal-Celigny et al, 2004) form the basis of prognostic assessments but these methods lack precision and more reliable biomarkers are needed (Gascoyne et al, 2010). Immunohistochemical studies have identified T-cell infiltration (Dave et al, 2004), in particular FOXP3 positive T regulatory (Treg) cells (Farinha et al, 2010), as being of prognostic significance. Tregs represent a subpopulation of T cells that act to suppress the immune responses of other cells, responsible for physiological prevention of excessive immune reactions and autoimmunity (Corthay, 2009), though altered numbers in tumours can influence survival, due in part to modulation of anti-tumoral immune response (Dranoff, 2005). Some studies report association of T cells with a poor outcome (Richendollar et al, 2011), though the majority of studies demonstrate association of T cells and Tregs with a favourable outcome (Carreras et al, 2006; Lee et al, 2006; Wahlin et al, 2007; Carreras et al, 2009; de Jong et al, 2009; Farinha et al, 2010; Gribben, 2010; Solal-Céligny et al, 2010; Wahlin et al, 2010; de Jong and Fest, 2011; Wahlin et al, 2011; Koch et al, 2012). Differences in prognostic significance exist between studies and this may in part be due to variable precision of T-cell subset identification, with several markers being required to identify particular types of T cells. For example, Richendollar et al (2011) demonstrated an unfavourable association for PD-1 positive T helper cells, whilst most favourable studies focus on Tregs (Wahlin et al, 2010). Several studies have also shown that the pattern of T cells is important for prognosis (Lee et al, 2006; de Jong et al, 2009; Farinha et al, 2010). There is therefore, (i) a need to co-localise several biomarkers/antigens in the same tissue section in order to identify tumour-infiltrating lymphocytes (TILs) more precisely, and (ii) a need to interpret patterns of TILs and to relate these patterns to prognosis. The first can be overcome by multiplex biomarker staining, as we have previously demonstrated (Tholouli et al, 2008). The second related but more subtle factor—the pattern of TILs positive for one or more biomarkers—is likely to be of biological and prognostic importance, as this in addition to the number of positive cells reflect biologically meaningful cellular interactions. Pattern identification using one or two biomarkers is feasible for an experienced pathologist but interpreting larger numbers (as expected with identification of increasingly complex immune cell interactions) is beyond unaided human interpretation. We have developed an automated prognostic feature/pattern-recognition tool, namely hypothesised interaction distribution (HID) analysis (Rose et al, 2013) to automatically and objectively identify prognostic patterns of different cells and biomarkers, and have tested this in FL. A diffuse pattern of FOXP3 positive cells has previously been shown to be associated with a favourable outcome in a subjective study using visual inspection and rating (Farinha et al, 2010) and in the present study we used this as a test scenario for automated prognostic feature identification, aiming to automatically and objectively determine, without prior knowledge, the presence/absence and type of prognostic pattern of FOXP3 and CD69 positive T cells in FL.
Materials and Methods
Patients and sample selection
Forty archived human lymph nodes from patients with a diagnosis of FL were obtained with informed consent from the archives of The Christie NHS Foundation Trust (TCFT), Manchester, UK. All biopsies were obtained at initial presentation prior to treatment and were routinely processed in 10% buffered formalin and paraffin embedded. Ethical permission for this study was granted by the Central Manchester Multicentre Research Ethical Committee (03/8/016). Diagnosis and grade in all cases were confirmed following consensus review by two pathologists (RJB and LPM). Where possible, FLIPI score was calculated using conventional criteria (Solal-Celigny et al, 2004).
Tissue microarray construction
Patient samples were analysed in a tissue microarray (TMA) to reduce reagent costs and facilitate imaging (Ilyas et al, 2013); we and others have used TMAs for analysis of FL in a wide range of studies, demonstrating validity of their use for this tumour type (Hedvat et al, 2002; De and Brown, 2010; Hipp et al, 2012; Sandison et al, 2013; Horn et al, 2014; McCarthy et al, 2014). TMA s were prepared using a Manual Tissue Arrayer (Beecher Instruments, Sun Prairie, WI, USA), each patient sample being represented in triplicate. Three cylindrical 1 mm diameter 100% tumour cores were taken from each paraffin-embedded FL sample and inserted into a recipient paraffin wax block.
Triplex immunohistochemistry
Triplex immunohistochemistry was performed for CD3, FOXP3 and CD69 on a Ventana Benchmark automated stainer (Ventana Systems, Oro Valley, AZ, USA) according to the manufacturer’s instructions and using the antibodies and detection reagents described in Table 1; a detailed description is given in Supplementary Information. Identification of TILs such as the Treg cells studied in this work require multiple staining for their identification, as a consequence of which CD3 was used in conjunction with FOXP3 and CD69 for multiplex T-cell staining required for TIL identification. T cells are of many subtypes as a consequence of which the pan T-cell marker CD3 alone is insufficient for identification of the Treg subset, necessitating additional use of the transcription factor FOXP3, which is a lineage-specific marker of the majority of Treg cells (Rudensky, 2011), together with CD69 which has been shown to identify a further subgroup of Tregs (Han et al, 2009).
Spectral imaging
A PerkinElmer Vectra (Hopkinton, MA, USA) multispectral slide scanner was used to capture image files at 10 nm intervals across the visible range (440 to 700 nm) and at × 200 magnification from which grey-scale distribution maps for CD3, FOXP3 and CD69 positivity for each core were derived by spectral un-mixing as previously described (Mansfield et al, 2008). Using these distribution maps, two multispectral image analyses were performed. Firstly Inform software (PerkinElmer, Hopkinton, MA, USA) was used to identify single positive CD3+ cells and double positive CD3/FOXP3+ and CD3/CD69+ cells, and the number and density of these measured in each core. Secondly Nuance software (PerkinElmer) was used to separately identify positive regions and their centroid co-ordinates for CD3, FOXP3 and CD69 positive staining, as required for HID analysis (Rose et al, 2013). The results of these two analyses were exported to Excel (Microsoft Corporation, Redmond, WA, USA) for use in Kaplan–Meier (K–M) survival and HID analyses, respectively, as detailed below.
K–M survival analysis
K–M survival analysis was performed using the number per core of either CD3 single positive, CD3/FOXP3 double positive or CD3/CD69 double positive cells, collected using Inform. For K–M analysis, the cell numbers were split at the 25th centile, the median and the 75th centile, such that quartile one was compared with quartiles two to four, quartiles one and two were compared with quartiles three and four, and quartiles one to three were compared with quartile four, respectively. All statistical analyses were performed using MedCalc version 11.4.40 (MedCalc Software, Ostend, Belgium).
Hypothesised interaction distance analysis
Spatial patterns of cells positive for FOXP3, CD69 or both were analysed using HID matrices (Rose et al, 2013). Briefly, a HID matrix is a two-dimensional statistical distribution (histogram) that records the absolute or relative frequencies of proximal pairs of regions (assumed to be individual cells) that are positive for each possible combination of marker (−+, +−, ++). A pair of regions was considered proximal if the regions were within an objectively chosen interaction distance of one another. A statistical approach was used to identify pairs of regions likely to correspond to individual cells; region centroid co-ordinates were collapsed into a single co-ordinate (Rose et al, 2013). Regions with areas greater than expected for single cells (200 pixels) were rejected. Absolute and relative frequency HID matrices were computed for each core. These were summarised using five scalar summary statistics (‘energy’ and maximum value of the absolute and relative frequencies, and the Shannon entropy of relative frequencies as detailed in Rose et al (2013); entropy is a measure of order of the arrangement of the cells. Each patient had multiple cores (i.e., multiple HID matrices) and so an average of each summary statistic was computed for each patient. A threshold can be applied to the average summary statistics to dichotomise patients into two groups; survival analysis was performed to determine if the groups had distinct survival functions and hence whether cell pattern is related to survival.
To estimate how well the approach would be able to predict whether an FL patient in the general population (as opposed to this sample) would have good vs poor survival, leave-one-out cross-validation was used to perform survival analyses and construct K–M curves. Each patient was left-out in turn and an optimal threshold was chosen automatically for each summary statistic using data from the left-in patients; for each summary statistic, its optimal threshold was used to assign the left-out patient to one of two survival groups (short or long survival). The null hypothesis of no difference in survival functions was tested for each summary statistic. HID analysis was performed with software written in-house using R version 2.13.0 (R-Core-Team, 2012).
To assess which cell pairs may be important in discriminating between FL survival groups, we used a random forest classifier (Breiman, 2001). Random forest classifiers can accurately model highly nonlinear relationships between multiple variables (here, the HID matrix entries) and their dependent classes—here, survival group, S (short) or L (long), as assigned using the entropy scalar summary. Random forest classifiers also maintain an internal estimate of the relative importance of each independent variable that is calculated using an internal cross-validation process. A random forest with 500 trees was built using version 4.6–7 of the randomForest R package (Liaw and Wiener, 2002). Cell pairings predictive of survival were identified as those associated with the best decrease in expected misclassification frequency (Gini index).
In addition, as entropy may be related to cell number and density, R^2 correlation (square of Pearson’s product–moment correlation coefficient) was performed against numbers and density of CD3, FOXP3 and CD69 positive cells with entropy value. Cox regression was performed using FLIPI, entropy score and tumour grade to determine whether entropy is predictive of outcome independent of FLIPI. To test the hypothesis that entropy is an independent predictor of survival, bivariate Cox regressions were performed for entropy against cell number or density for CD3, FOXP3 and CD69; single multivariate Cox regression for entropy against cell number or density was not performed owing to small sample size.
Results
Patient samples
A total of 40 patient samples had adequate tissue cores in the TMA and full clinical follow-up to death or to a maximum of 171 months (24 male, 16 female, median age 55.5 years at diagnosis, range 34–75 years). At the time of analysis, 29 patients had died and 11 were alive (median survival 56 months, range 8–171 months); the cohort of patients was treated at The Christie Hospital prior to the use of Rituximab and had a higher than usual rate of transformation reflective of referral to a cancer centre, resulting in relatively poor overall survival compared with present outcomes. Patient demographics including tumour grade and FLIPI are detailed in Table 2.
Triplex immunohistochemistry
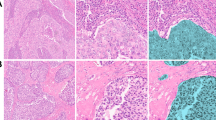
Triplex immunohistochemistry was successfully performed for CD3, FOXP3 and CD69, with an expected pattern and cellular localisation of positive staining. Representative images of triplex staining and grey-scale spectrally unmixed positive-distribution maps for CD3, FOXP3 and CD69 are shown in Figure 1.
Triplex immunohistochemistry for CD3, FOXP3 and CD69. Representative images of (A) triplex immunohistochemistry for CD3, FOXP3 and CD69, and of grey scale displayed spectrally unmixed positive-distribution maps/images for (B) CD3, (C) FOXP3 and (D) CD69; all images: magnification × 20, imaged using a Perkin-Elmer Vectra multispectral imaging system and acquired and processed using Perkin-Elmer Inform and Nuance software.
K–M survival analysis demonstrated favourable outcome with higher numbers of CD3+, FOXP3+/CD3+ and CD69+/CD3+ cells
Higher numbers of CD3 single positive cells were significantly associated with a favourable outcome by K–M analysis for cell numbers whether split at the lower quartile (P=0.0072), the median (P=0.0001) or the upper quartile (P=0.0011; Figure 2A).
(A) Association of CD3 single positive cells with outcome. K–M survival analysis based on numbers of CD3 single positive cells. Analyses are shown using either (i) the 25th centile, (ii) median or (iii) 75th centile to split the patients into two groups. Higher numbers of CD3 single positive cells were significantly associated with favourable outcome with patients split at the 25th centile (P=0.0072), the median (P=0.0001) or the 75th centile (P=0.0011). For each K–M plot survival probability as a percentage is plotted on the vertical axis and survival time in months is on the horizontal axis. (B) Association of FOP3/CD3 double positive cells with outcome. K–M survival analysis based on numbers of FOXP3/CD3 double positive cells. Analyses are shown using either (i) the 25th centile, (ii) median or (iii) 75th centile to split the patients into two groups. Higher numbers of FOP3/CD3 double positive cells were significantly associated with favourable outcome with patients split at the 25th centile (P=0.0013), the median (P=0.001) or the 75th centile (P=0.002). For each K–M plot survival probability as a percentage is plotted on the vertical axis and survival time in months is on the horizontal axis. (C) Association of CD69/CD3 double positive cells with outcome. K–M survival analysis based on numbers of CD69/CD3 double positive cells. Analyses are shown using either (i) the 25th centile, (ii) median or (iii) 75th centile to split the patients into two groups. Higher numbers of CD69/CD3 double positive cells were significantly associated with favourable outcome with patients split at the 25th centile (P=0.9925), the median (P=0.0152) or the 75th centile (P=0.0006). For each K–M plot survival probability as a percentage is plotted on the vertical axis and survival time in months is on the horizontal axis.
Higher numbers of FOXP3/CD3 double positive cells were also significantly associated with a favourable outcome by K–M analysis for cell numbers, whether split at the lower quartile (P=0.0232), the median (P=0.004) or the upper quartile (P=0.0031; Figure 2B).
Higher numbers of CD69/CD3 double positive cells were significantly associated with a favourable outcome by K–M analysis for cell numbers split at the median (P=0.0152) and at the upper quartile (P=0.0006; Figure 2C).
Hypothesised interaction distance analysis demonstrated association of favourable outcome with a high entropy/diffuse pattern of FOXP3 positive and CD69 positive cells
Each of the five summary statistics was significantly associated with a difference in survival, as shown in Table 3a. For each summary statistic, Table 3a presents the median value of the summary statistic in each survival group, the median survival time of each group, the difference in median survival between each group and the P-value from the survival analysis. The P-values have not been corrected for multiple comparisons, but all except that for energy computed for relative frequencies would survive Bonferroni’s correction at a significance level of 0.05.
For the entropy summary statistic in particular, patients with a higher entropy value had significantly better outcome (P=0.00255) than patients with low entropy, with a median survival of 142 months compared with a median survival of 35.5 months (Figure 3A). High entropy represents a more dispersed/diffuse pattern of T cells compared with those patients with lower entropy, where the pattern of T cells was more aggregated. Representative core images are shown for patients with high entropy/favourable outcome and low entropy/unfavourable outcomes in Figure 3B and C.
Higher Shannon entropy, representing a more diffuse pattern of FOXP3 and CD69 positivity, is associated with favourable outcome. (A) K–M analysis for Shannon entropy calculated by HID analysis, demonstrating association of higher entropy level, above the median, with favourable outcome (P=0.00255). Representative images of patient samples with (B) high Shannon entropy, demonstrating diffuse pattern of FOXP3 positive cells and (C) low Shannon entropy, demonstrating an aggregated pattern of FOXP3 positive cells; magnification: × 20.
Only 5.66% of patients were misclassified using the random forest. Inspecting the relative importance estimates for the random forest suggested that all entries of the HID matrices carry information that allows patients with short vs long survival to be discriminated, as shown in Table 3b for two typical patients, one with short survival, another with long survival. Therefore the spatial pattern of FOXP3 and CD69 positive cells is likely predictive of survival. However, three types of cell pairs had particularly large values of relative importance. In order of decreasing relative importance these were as follows: (i) both cells in the pair positive for FOXP3 only, (ii) both cells in the pair positive for CD69 only and (iii) pairs with one cell positive for FOXP3 only and the other for CD69 only. Examples of the way in which patients with short and long survival differ in terms of these cell pairings is shown in Table 3b. The most commonly occurring cell pairing in all patients (i.e., the mode of the HID matrices) was for cell pairs where both cells in the pair were positive for FOXP3 only.
Both the number and density of CD3, FOXP3 and CD69 positive cells were correlated with entropy, that is, higher entropy/dispersed pattern was associated with higher number and density of cells for all the three markers except FOXP3 density (CD3 number P=0.0002, CD3 density P=0.0007, FOXP3 number P=0.0317, FOXP3 density P=0.0692, CD69 number P<0.0001, CD69 density P<0.0001; Table 4a).
Entropy score (high or low) was not associated with FLIPI score (P=0.3696) or grade (P=0.1923). Cox regression demonstrated lack of significance for FLIPI (P=0.2219), entropy (P=0.2486) and grade (P=0.6183). Bivariate Cox regression analysis demonstrated significance for entropy in the models including number of FOXP3 cells and density of FOXP3 cells (Table 4b). In both these models the estimate of hazard ratio for entropy was less than one, indicating that higher entropy is associated with longer survival. The two models in which the non-entropy variable was significant were those for CD3 cell number and CD3 cell density though for both of these models the estimate of hazard ratio for CD3 number or density was essentially equal to one, and the very narrow confidence interval may indicate that those results were unreliable. These results indicate that following control for entropy in bivariate Cox regressions, entropy was the only variable useful for explaining survival.
Discussion
Histopathological diagnosis relies on the visual study of cell morphology, often with additional immunohistochemical staining, usually for just one or two markers at a time. Increasingly, personalised therapy is creating a need for multiple biomarkers to enable more precise characterisation of cancer cells and facilitate theragnosis. This is true for TILs, which are important in novel immunological therapies and require multiple markers for their identification. Cells positive for any given number of biomarkers can be enumerated and quantified but there are currently no commercially available methods for objectively analysing the spatial pattern of cells positive for multiple biomarkers. This is an important requirement as patterns are likely to reflect cellular interactions and, in turn, cellular behaviour and clinical outcomes. We have developed an automated method, HID analysis, for analysing the spatial patterns of multiple cellular markers, using multispectral imaging and HID analysis (Rose et al, 2013).
The present study used this approach in automated and objective prognostic feature identification to determine, without prior knowledge, the presence or absence of a prognostic pattern for FOXP3 and CD69 positive T cells in FL. HID matrices and hence entropy values were constructed as previously described (Rose et al, 2013). Briefly, the pairing distance (the distance within which a pair of regions positive for distinct stains are assumed to correspond to the same cell) and the interaction distance (the distance within which regions assumed to be distinct cells are assumed to be able to interact), are chosen objectively via a statistical optimisation that is performed across the patient cohort. As stated in Rose et al (2013), for full objectivity these distances would ideally be computed within an overarching leave-one-out framework (or similar), but this is not computationally tractable.
Higher numbers of CD3 single positive, FOXP3/CD3 double positive and CD69/CD3 double positive cells were significantly associated with a favourable outcome by K–M analysis whilst HID analysis demonstrated a significant association of cellular pattern with favourable outcome, for example, with higher entropy being associated with longer median survival (142 months) compared with those with lower entropy (35.5 months; P=0.00255). Because higher entropy reflects a more diffuse cellular pattern, a diffuse pattern of FOXP3 and CD69 positive T cells was therefore associated with favourable outcome. These results demonstrate the ability of HID analysis to automatically and objectively identify the prognostic patterns of cellular distribution. We have demonstrated that a diffuse pattern of FOXP3 and CD69 cells is associated with a favourable outcome in FL, replicating, in an automated and objective manner, the subjective finding by Farinha et al (2010) that a diffuse pattern of FOXP3 positive Tregs is a favourable prognostic feature. Entropy was not significantly associated with either FLIPI score or tumour grade, but was inversely correlated with the number and density of CD3, FOXP3 and CD69 positive cells (CD3 number P=0.0003, CD3 density P=0.0015, FOXP3 number P=0.0010, FOXP3 density P=0.0035, CD69 number P<0.0001, CD69 density P<0.0001).
Many previous studies have identified T-cell infiltration (Dave et al, 2004), in particular FOXP3 positive Treg cells (Farinha et al, 2010), as being of prognostic significance in FL. Most demonstrate an association of T cells and Tregs with favourable outcome (Carreras et al, 2006; Lee et al, 2006; Wahlin et al, 2007; Carreras et al, 2009; de Jong et al, 2009; Farinha et al, 2010; Gribben, 2010; Solal-Céligny et al, 2010; Wahlin et al, 2010; de Jong and Fest, 2011; Wahlin et al, 2011; Koch et al, 2012) although some report an unfavourable outcome (Richendollar et al, 2011). Discrepancy between studies may be owing to differences in precision of T-cell subset identification, or in pattern analysis, the pattern of T cells being important for prognosis (Lee et al, 2006; de Jong et al, 2009; Farinha et al, 2010), whilst the present study concords with the majority of previous studies in which higher levels of T cells are associated with longer survival in FL (Dave et al, 2004).
Although there is a large body of evidence supporting a favourable prognostic role for T cells in general, Tregs, which are a subset of CD4+ cells, are of particular importance. Farinha et al (2010) demonstrated a favourable association of a diffuse pattern of FOXP3 positive Tregs, whilst a follicular or peri-follicular pattern was unfavourable. Interestingly, and consequent upon their role in immune tolerance, higher numbers of FOXP3 Tregs are associated with a poor outcome in some non-haematological cancers (deLeeuw et al, 2012), whilst cases of adult T-cell leukaemia/lymphoma expressing higher levels of FOXP3 also have a worse prognosis (Roncador et al, 2005; Marzano et al, 2009). Conversely studies in FL have reported a positive association between Tregs and overall survival (Carreras et al, 2006; Farinha et al, 2010), which the present study supports. Available evidence indicates that the increase of Tregs seen in haematological malignancies is because of a proliferative response and not owing to the disruption of senescence suggesting a tumour-specific recruitment. However, the exact mechanisms responsible for their effects in FL remain unknown. Tregs have two suppressive mechanisms involving direct cell–cell contact and cytokine production (Mougiakakos et al, 2010). The deployed mechanism ultimately depends on the nature of the tumour micro-environment. Immunosuppressive cytokines (IL-10 and TGF-β) secreted by Tregs suppress tumour-promoting mechanisms and/or induce tumour cell death (Whiteside, 2010) and may thus represent an explanation for the relationship between increased number of Tregs and improved patient survival.
It is known that Tregs are involved in immune tolerance and act to suppress the activity of T cells, B cells and antigen presenting cells, preventing self-reactivity. FOXP3 is thought to have a role in this via binding to the promotors of genes involved in Treg function, with repression of gene transcription following T-cell receptor stimulation (Marson et al, 2007). The specific dynamics of FOXP3+ Treg interaction with malignant B cells is therefore important in determining outcome. Reactive lymph nodes contain FOXP3+ Tregs at the interface of the T cell and B cell areas, where they act to suppress CD4+ T cells, and they are increased in FL compared with reactive nodes (Lim et al, 2005). They also act to induce the pro-tumourigenic M2 macrophage phenotype (Tiemessen et al, 2007) and, where they have a follicular pattern, are associated with ineffective tumour immunity. This allows FL cells to avoid immunosurveillance, resulting in inferior survival and increased risk of transformation (Farinha et al, 2010). Enrichment of Tregs in the follicles directly interacting with the tumour cells would therefore clearly contribute to a poorer outcome in FL. Tregs are activated in the T-cell zone outside of the follicle, exerting their immunosuppressive effect upon migration to the follicles where they interact with the centrocytes (Linterman et al, 2012). Our finding of poor outcome in patients with a more aggregated pattern may therefore represent a higher degree of interaction between Tregs and malignant B cells, with greater pro-tumoural immunity, underlining the importance of the pattern of cellular interaction.
Farinha et al (2010) demonstrated prognostic importance of FOXP3 Tregs using expert morphological assessment, which is difficult to determine ‘by eye’ for more than one and certainly not more than two markers. Figure 4 illustrates two subtly different distributions; one, in panel 4A, is more diffuse and of higher entropy, compared with another, in panel 4B, which is more aggregated and of lower entropy. Distinction of these patterns by eye is difficult and difficulty increases further as the number of markers rise above three. HID analysis is able to objectively and automatically quantitate the different patterns, enabling them to be used in statistical analyses, in order to identify prognostic patterns associated with favourable or unfavourable outcome. This is illustrated in Figure 4; panels c and d show representative patient samples with diffuse and aggregated cells positive for FOXP3 and CD69, respectively, in turn representing high- and low-entropy patterns with favourable and unfavourable outcomes. The method used in the present study enabled prognostic pattern to be identified automatically using three markers, CD3, CD69 and FOXP3, and is theoretically scalable to larger number of markers. Furthermore, the data in Table 3b indicate that the example patient with poor survival lacks proximity for cells that are positive for CD69 (either alone or in combination with FOXP3) to cells that are positive for FOXP3, and that CD69 positive cells are likely to be having a regulatory effect on the tumor cells in terms of the driver of disease progression (transformation, proliferation, etc.). Further, this can only be determined with an analysis such as HID analysis that considers the spatial heterogeneity of distinct cellular subtypes.
Overview of method taken to identify prognostic patterns of multiple markers illustrating the ability to automatically and objectively distinguish between subtly different patterns. (A) and (B) represent cellular patterns for three cellular phenotypes. The pattern in (A) is slightly more diffuse than that in (B) but this is difficult to identify by eye, such that an attempt by eye to determine the association of a range of such variable patterns with patient outcome would not be feasible reliably. HID analysis quantifies the nature of the cellular pattern such that metrics defining the pattern, such as Shannon entropy, can be used to objectively test the association of pattern with outcome, using K–M analysis. This can objectively demonstrate different outcomes for the distributions of cells in (A) and (B), which correspond to diffuse or aggregated cellular patterns in tissue samples, as shown at the bottom left of (C) and (D), respectively.
Although it is true that higher entropy is associated with higher numbers of T cells and Tregs, bivariate Cox regression analysis demonstrated, once controlled for entropy, that entropy is predictive of outcome independent of cell number or density. Furthermore, though there were significant correlations between entropy and cell number and density the R^2 values for these correlations were all relatively low (Table 4a), ranging from 0.1026 to 0.3665 for all except CD69. Consequently entropy is able to describe an additional feature associated with survival, namely pattern, either dispersed or aggregated.
Automation will increasingly be needed for multiple biomarker analysis as part of translational research. However, whilst automated cell counting is now routine (Ali et al, 2013), quantitative, automated measurement of cellular spatial distribution pattern as opposed to pure cell numbers has rarely been achieved (Beck et al, 2011; Rimm, 2011; Yuan et al, 2012). Setiadi et al (2010) used a statistic known as the L function, a variant of Ripley’s K function, to quantify and identify statistically significant differences in spatial grouping of B and T cells in healthy lymph nodes and cancer draining lymph nodes in breast cancer patients showing a lack of B-cell localisation in the extrafollicular region of the tumour draining lymph nodes. However, the approach used was unable to correlate cellular pattern with outcome as in the present study, as the L function does not yield an outcome statistic usable in K–M survival analysis.
In conclusion, the spatial pattern of cells is important for their biological behaviour and will therefore be important for prognosis of tumours. Ability to identify such prognostic patterns ‘by eye’ will become increasingly difficult as the number of biomarkers increase. The present study used an objective and quantitative method to automatically identify prognostic patterns of Tregs in FL, albeit in a relatively small sample size. The results therefore require additional validation in large, independent sample sets but provide evidence of confidence for doing such larger studies. Although such patterns have been identified before, their identification has relied on prior knowledge and has been manual. Our approach removes the possibility for investigator bias and is theoretically applicable to large numbers of biomarkers, representing a novel method for translational and diagnostic pathology and theragnostics.
Change history
20 October 2015
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Ali HR, Irwin M, Morris L, Dawson SJ, Blows FM, Provenzano E, Mahler-Araujo B, Pharoah PD, Walton NA, Brenton JD, Caldas C (2013) Astronomical algorithms for automated analysis of tissue protein expression in breast cancer. Br J Cancer 108: 602–612.
Beck AH, Sangoi AR, Leung S, Marinelli RJ, Nielsen TO, van de Vijver MJ, West RB, van de Rijn M, Koller D (2011) Systematic analysis of breast cancer morphology uncovers stromal features associated with survival. Sci Transl Med 3: 108ra113.
Breiman L (2001) Random forests. Mach Learn 45: 27.
Carreras J, Lopez-Guillermo A, Fox BC, Colomo L, Martinez A, Roncador G, Montserrat E, Campo E, Banham AH (2006) High numbers of tumor-infiltrating FOXP3-positive regulatory T cells are associated with improved overall survival in follicular lymphoma. Blood 108: 2957–2964.
Carreras J, Lopez-Guillermo A, Roncador G, Villamor N, Colomo L, Martinez A, Hamoudi R, Howat WJ, Montserrat E, Campo E (2009) High numbers of tumor-infiltrating programmed cell death 1-positive regulatory lymphocytes are associated with improved overall survival in follicular lymphoma. J Clin Oncol 27: 1470–1476.
Corthay A (2009) How do regulatory T cells work? Scand J Immunol 70: 11.
Dave SS, Wright G, Tan B, Rosenwald A, Gascoyne RD, Chan WC, Fisher RI, Braziel RM, Rimsza LM, Grogan TM, Miller TP, LeBlanc M, Greiner TC, Weisenburger DD, Lynch JC, Vose J, Armitage JO, Smeland EB, Kvaloy S, Holte H, Delabie J, Connors JM, Lansdorp PM, Ouyang Q, Lister TA, Davies AJ, Norton AJ, Muller-Hermelink HK, Ott G, Campo E, Montserrat E, Wilson WH, Jaffe ES, Simon R, Yang L, Powell J, Zhao H, Goldschmidt N, Chiorazzi M, Staudt LM (2004) Prediction of survival in follicular lymphoma based on molecular features of tumor-infiltrating immune cells. N Engl J Med 351: 2159–2169.
De J, Brown RE (2010) Tissue-microarray based immunohistochemical analysis of survival pathways in nodular sclerosing classical Hodgkin lymphoma as compared with non-Hodgkin's lymphoma. Int J Clin Exp Med 3: 55–68.
de Jong D, Fest T (2011) The microenvironment in follicular lymphoma. Best Pract Res Clin Haematol 24: 135–146.
de Jong D, Koster A, Hagenbeek A, Raemaekers J, Veldhuizen D, Heisterkamp S, de Boer JP, van Glabbeke M (2009) Impact of the tumor microenvironment on prognosis in follicular lymphoma is dependent on specific treatment protocols. Haematologica 94: 70–77.
deLeeuw RJ, Kost SE, Kakal JA, Nelson BH (2012) The prognostic value of FoxP3+ tumor-infiltrating lymphocytes in cancer: a critical review of the literature. Clin Cancer Res 18: 3022–3029.
Dranoff G (2005) The therapeutic implications of intratumoral regulatory T cells. Clin Cancer Res 11: 8226–8229.
Farinha P, Al-Tourah A, Gill K, Klasa R, Connors JM, Gascoyne RD (2010) The architectural pattern of FOXP3-positive T cells in follicular lymphoma is an independent predictor of survival and histologic transformation. Blood 115: 289–295.
Freedman A (2012) Follicular lymphoma: 2012 update on diagnosis and management. Am J Hematol 87: 988–995.
Gascoyne RD, Rosenwald A, Poppema S, Lenz G (2010) Prognostic biomarkers in malignant lymphomas. Leuk Lymphoma 51 (Suppl 1): 11–19.
Gribben JG (2010) Implications of the tumor microenvironment on survival and disease response in follicular lymphoma. Curr Opin Oncol 22: 424–430.
Han Y, Guo Q, Zhang M, Chen Z, Caio X (2009) CD69+ CD4+ CD25− T cells, a new subset of regulatory T cells, suppress T cell proliferation through membrane-bound TGF-beta 1. J Immunol 182: 10.
Hedvat CV, Hegde A, Chaganti RS, Chen B, Qin J, Filippa DA, Nimer SD, Teruya-Feldstein J (2002) Application of tissue microarray technology to the study of non-Hodgkin's and Hodgkin's lymphoma. Hum Pathol 33: 968–974.
Hipp JA, Hipp JD, Lim M, Sharma G, Smith LB, Hewitt SM, Balis UG (2012) Image microarrays derived from tissue microarrays (IMA-TMA): New resource for computer-aided diagnostic algorithm development. J Pathol Inform 3: 24.
Horn H, Bausinger J, Staiger AM, Sohn M, Schmelter C, Gruber K, Kalla C, Ott MM, Rosenwald A, Ott G (2014) Numerical and structural genomic aberrations are reliably detectable in tissue microarrays of formalin-fixed paraffin-embedded tumor samples by fluorescence in-situ hybridization. PLoS One 9: e95047.
Ilyas M, Grabsch H, Ellis IO, Womack C, Brown R, Berney D, Fennell D, Salto-Tellez M, Jenkins M, Landberg G, Byers R, Treanor D, Harrison D, Green AR, Ball G, Hamilton P (2013) Guidelines and considerations for conducting experiments using tissue microarrays. Histopathology 62: 827–839.
Koch K, Hoster E, Unterhalt M, Ott G, Rosenwald A, Hansmann ML, Engelhard M, Hiddemann W, Klapper W (2012) The composition of the microenvironment in follicular lymphoma is associated with the stage of the disease. Hum Pathol 43: 8.
Lee AM, Clear AJ, Calaminici M, Davies AJ, Jordan S, MacDougall F, Matthews J, Norton AJ, Gribben JG, Lister TA, Goff LK (2006) Number of CD4+ cells and location of forkhead box protein P3-positive cells in diagnostic follicular lymphoma tissue microarrays correlates with outcome. J Clin Oncol 24: 5052–5059.
Liaw A, Wiener M (2002) Classification and regression by randomForest. R News 2: 5.
Lim HW, Hillsamer P, Banham AH, Kim CH (2005) Cutting edge: direct suppression of B cells by CD4+ CD25+ regulatory T cells. J Immunol 175: 4180–4183.
Linterman MA, Liston A, Vinuesa CG (2012) T-follicular helper cell differentiation and the co-option of this pathway by non-helper cells. Immunol Rev 247: 143–159.
Mansfield JR, Hoyt C, Levenson RM (2008) Visualization of microscopy-based spectral imaging data from multi-label tissue sections. Curr Protoc Mol Biol Chapter 14: Unit 14 19.
Marson A, Kretschmer K, Frampton GM, Jacobsen ES, Polansky JK, MacIsaac KD, Levine SS, Fraenkel E, von Boehmer H, Young RA (2007) Foxp3 occupancy and regulation of key target genes during T-cell stimulation. Nature 445: 931–935.
Marzano AV, Vezzoli P, Fanoni D, Venegoni L, Berti E (2009) Primary cutaneous T-cell lymphoma expressing FOXP3: a case report supporting the existence of malignancies of regulatory T cells. J Am Acad Dermatol 61: 348–355.
McCarthy A, Marzec J, Clear A, Petty RD, Coutinho R, Matthews J, Wilson A, Iqbal S, Calaminici M, Gribben JG, Jia L (2014) Dysregulation of autophagy in human follicular lymphoma is independent of overexpression of BCL-2. Oncotarget 5: 11653–11668.
Mougiakakos D, Choudhury A, Lladser A, Kiessling R, Johansson CC (2010) Regulatory T cells in cancer. Adv Cancer Res 107: 57–117.
R-Core-Team (2012) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria.
Richendollar BG, Pohlman B, Elson P, Hsi ED (2011) Follicular programmed death 1-positive lymphocytes in the tumor microenvironment are an independent prognostic factor in follicular lymphoma. Hum Pathol 42: 552–557.
Rimm DL (2011) C-path: a Watson-like visit to the pathology lab. Sci Transl Med 3: 108fs8.
Roncador G, Garcia JF, Maestre L, Lucas E, Menarguez J, Ohshima K, Nakamura S, Banham AH, Piris MA (2005) FOXP3, a selective marker for a subset of adult T-cell leukaemia/lymphoma. Leukemia 19: 2247–2253.
Rose CJ, Naidoo K, Clay V, Linton K, Radford JA, Byers RJ (2013) A statistical framework for analyzing hypothesized interactions between cells imaged using multispectral microscopy and multiple immunohistochemical markers. J Pathol Inform 4: S4.
Rudensky AY (2011) Regulatory T cells and Foxp3. Immunol Rev 241: 260–268.
Sandison HE, Usher S, Karimiani EG, Ashton G, Menasce LP, Radford JA, Linton K, Byers RJ (2013) PLK1 and YY1 interaction in follicular lymphoma is associated with unfavourable outcome. J Clin Pathol 66: 764–767.
Setiadi AF, Ray NC, Kohrt HE, Kapelner A, Carcamo-Cavazos V, Levic EB, Yadegarynia S, van der Loos CM, Schwartz EJ, Holmes S, Lee PP (2010) Quantitative, architectural analysis of immune cell subsets in tumor-draining lymph nodes from breast cancer patients and healthy lymph nodes. PLoS One 5: e12420.
Solal-Céligny P, Cahu X, Cartron G (2010) Follicular lymphoma prognostic factors in the modern era: what is clinically meaningful? Int J Hematol 92: 9.
Solal-Celigny P, Roy P, Colombat P, White J, Armitage JO, Arranz-Saez R, Au WY, Bellei M, Brice P, Caballero D, Coiffier B, Conde-Garcia E, Doyen C, Federico M, Fisher RI, Garcia-Conde JF, Guglielmi C, Hagenbeek A, Haioun C, LeBlanc M, Lister AT, Lopez-Guillermo A, McLaughlin P, Milpied N, Morel P, Mounier N, Proctor SJ, Rohatiner A, Smith P, Soubeyran P, Tilly H, Vitolo U, Zinzani PL, Zucca E, Montserrat E (2004) Follicular lymphoma international prognostic index. Blood 104: 1258–1265.
Tholouli E, Sweeney E, Barrow E, Clay V, Hoyland JA, Byers RJ (2008) Quantum dots light up pathology. J Pathol 216: 275–285.
Tiemessen MM, Jagger AL, Evans HG, van Herwijnen MJ, John S, Taams LS (2007) CD4+CD25+Foxp3+ regulatory T cells induce alternative activation of human monocytes/macrophages. Proc Natl Acad Sci USA 104: 19446–19451.
Wahlin BE, Aggarwal M, Montes-Moreno S, Gonzalez LF, Roncador G, Sanchez-Verde L, Christensson B, Sander B, Kimby E (2010) A unifying microenvironment model in follicular lymphoma: outcome is predicted by programmed death-1–positive, regulatory, cytotoxic, and helper T cells and macrophages. Clin Cancer Res 16: 637–650.
Wahlin BE, Sander B, Christensson B, Kimby E (2007) CD8+ T-cell content in diagnostic lymph nodes measured by flow cytometry is a predictor of survival in follicular lymphoma. Clin Cancer Res 13: 388–397.
Wahlin BE, Sundstrom C, Holte H, Hagberg H, Erlanson M, Nilsson-Ehle H, Linden O, Nordstrom M, Ostenstad B, Geisler CH, Brown Pde N, Lehtinen T, Maisenholder M, Tierens AM, Sander B, Christensson B, Kimby E (2011) T cells in tumors and blood predict outcome in follicular lymphoma treated with rituximab. Clin Cancer Res 17: 4136–4144.
Whiteside TL (2010) Immune responses to malignancies. J Allergy Clin Immunol 125: S272–S283.
Yuan Y, Failmezger H, Rueda OM, Ali HR, Graf S, Chin SF, Schwarz RF, Curtis C, Dunning MJ, Bardwell H, Johnson N, Doyle S, Turashvili G, Provenzano E, Aparicio S, Caldas C, Markowetz F (2012) Quantitative image analysis of cellular heterogeneity in breast tumors complements genomic profiling. Sci Transl Med 4: 157ra143.
Acknowledgements
LSN was supported by an Undergraduate Intercalated BSc award from the British Division of the International Academy of Pathology. CJR was funded by a joint AstraZeneca/University of Manchester Fellowship.
Author contributions
LSN performed the research, collected, analysed and interpreted all data and wrote the manuscript. JRM and RL performed the research, analysed the data and wrote the manuscript. KO analysed the data and wrote the manuscript. LPM and KML provided clinical liaison and wrote the manuscript. CJR and RJB designed the research, analysed and interpreted all data and wrote the manuscript. ZS collected and analysed data. All authors contributed to research design, or acquisition, analysis and interpretation of data. All authors contributed to drafting and editing the manuscript and all approved the final version submitted.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
Supplementary Information accompanies this paper on British Journal of Cancer website
Supplementary information
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Nelson, L., Mansfield, J., Lloyd, R. et al. Automated prognostic pattern detection shows favourable diffuse pattern of FOXP3+ Tregs in follicular lymphoma. Br J Cancer 113, 1197–1205 (2015). https://doi.org/10.1038/bjc.2015.291
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2015.291
Keywords
This article is cited by
-
Immune infiltrate diversity confers a good prognosis in follicular lymphoma
Cancer Immunology, Immunotherapy (2021)
-
New developments in the pathology of malignant lymphoma. A review of the literature published from September 2015–December 2015
Journal of Hematopathology (2016)