Abstract
In chronic lymphocytic leukemia (CLL), the detection of minimal residual disease (MRD) correlates with outcome in the trial setting. However, MRD assessment does not guide routine clinical management and its assessment remains complex. We incorporated detection of the B cell, tumor-specific antigen CD160 to develop a single-tube, flow cytometry assay (CD160FCA) for CLL MRD to a threshold of 10−4 to 10−5. One hundred and eighty-seven patients treated for CLL were enrolled. Utilizing the CD160FCA methodology, there was a high level of comparison between blood and bone marrow (R=0.87, P<0.001). In a validation cohort, CD160FCA and the international standardised approach of the European Research Initiative on CLL group demonstrated high concordance (R=0.91, P<0.01). Patients in complete remission (CR) and CD160FCA negative had longer event-free survival (EFS) (63 vs 16 months, P<0.01) and prolonged time to next treatment (60 vs 15 months, P<0.001) vs MRD positive patients; with a median time to MRD positivity of 36 months. In multivariate analysis, CD160FCA MRD detection was independently predictive of EFS in patients in CR and even predicted EFS in the good-risk cytogenetic subgroup. CD160FCA offers a simple assay for MRD detection in CLL and gives prognostic information across different CLL risk groups.
Similar content being viewed by others
Introduction
The concept of minimal residual disease (MRD) assessment and eradication in chronic lymphocytic leukemia (CLL) is still considered a novel approach to patient care. Historically, the investigation of MRD was not clinically relevant as treatment protocols were designed with palliation in mind, and stabilising symptoms with alkylating agents and complete remission (CR) rates were low.1, 2, 3 However, newer chemo-immunotherapeutic regimens have achieved CR rates of 60–70%, and have evaluated MRD in this context.4, 5 Nevertheless, relapse is inevitable after an initial CR due to low-level residual cells. CR is currently defined as the absence of constitutional symptoms and organomegaly, absolute neutrophil count ⩾1500 per microlitre, a platelet count >100 × 109/l and a hemoglobin level >11.0 g/dl; the bone marrow (BM) must be normocellular with <30% lymphocytes.6 MRD assessment can stratify patients in CR into MRD-positive and -negative groups, with MRD-negative patients having a survival advantage.4, 7, 8, 9
MRD assessment in CLL can be demonstrated using either flow cytometry or allele-specific polymerase chain reaction (PCR).10, 11, 12 Although allele-specific PCR has a high sensitivity (1 in 106), it is complex, expensive and a pre-treatment sample is a prerequisite. The European Research Initiative on CLL have proposed an international standardised flow cytometric approach (ISA) and compared it against real-time quantitative allele-specific oligonucleotide immunoglobulin heavy chain gene PCR.10 The authors demonstrated concordant results between flow cytometry and real-time quantitative allele-specific oligonucleotide immunoglobulin heavy chain gene PCR in 94.7% of cases.10, 11 Peripheral blood (PB) analysis was at least as sensitive as BM for MRD assessment, with the exception of treatment with Alemtuzumab, where BM samples were necessary.
The recent description of the restricted expression in the B-cell lineage of the cell surface marker CD160 to malignant, but not normal B-cells, makes it an ideal candidate marker for MRD detection.13 In normal lymphocytes, CD160 is expressed on the majority of circulating NK cells, a subset of cytotoxic T cells, but not B cells.14, 15 In CLL, CD160 mediates PI3K-dependent cellular activation: upregulation of Bcl-2, Bcl-XL, improved in vitro survival and cytokine secretion.16
There is near-universal expression of CD160 in CLL, monoclonal B-cell lymphocytosis and hairy cell leukemia, which was established using the CD160 flow cytometric assay (CD160FCA).13 In this report, we have investigated the utility of CD160FCA for MRD detection in CLL and compared it with the current recommended standard.10
Patients and methods
CD160 flow cytometric assay for MRD detection
CD160FCA is a six-color assay, which has been slightly modified since previously reported, but still uses the same whole-blood analysis protocol and simple gating strategy that has been modified by one additional CD45 vs Side Scatter gate since previously reported.13 The CD160FCA incorporates CD2-FITC (Clone S5.2), CD5-APC (Clone L17F12), CD19-PerCPcy5.5 (Clone SJ25C1), CD23-APC (Clone EBVCS-5) and CD45 V500 (Clone Hi30) (BD Biosciences, Oxford, UK); CD160-PE (Clone BY55; IgM isotype; Immunotech, Beckman Coulter, Marseilles, France). All monoclonal antibodies used in the study underwent titration to determine the optimum concentration of monoclonal antibody for a given number of cells. To determine the optimal saturating concentration, a serial doubling dilution of the fluorochrome-labeled antibody was tested over a range above and below that of the manufacturer's recommendations. The intrinsic spectral overlap of the different fluorochromes in the CD160FCA was corrected using compensation matrices and automatic compensation beads (BD Biosciences). The CD160FCA underwent steric hindrance analysis to assess if antigen expression was reduced or absent if the antibodies were binding the same macromolecular complex. Further evaluation for reagent interaction altering assay performance was efficiently assessed by comparing the median fluorescence intensity of a sample prepared with all antibodies of interest with a series of preparations in which each lacks one of the component fluorochrome-conjugated monoclonal antibodies.
Where possible 500 000 events were acquired for each MRD patient on a FACS Canto II with standard laser and filter configuration (BD Biosciences). Daily calibration of the instrument was performed using CS&T beads (BD Biosciences). Data were acquired and analyzed by means of BD FACS Diva clinical software (version 6.1.3) for enhanced acquisition analysis, which determined the median fluorescence intensity of CD160 on the CD2negCD5+CD19+CD23+ population. Residual disease was based on the number of events co-expressing CD2negCD5+CD19+CD23+CD160+ out of the total number of events collected.
Simulation of MRD
To assess the utility of CD160 in the monitoring of MRD and the possible limit of detection, samples simulating MRD specimens were prepared by mixing separated untreated CLL cells (n=5) with normal leukocytes (whole blood) from donors. Serial dilutions were performed from 1:1–1:100 000 to determine the sensitivity and specificity of the assay.
CD160 PCR analysis
Qualitative MRD assessment was performed on total RNA extracted from freshly isolated CD19+ B-cells as previously described.13 The specific primers for complementary DNA synthesis were designed on published sequences from human CD160 (NM_007053): BY-TM (5′-GCCAGAAGCCAGAAGTCAGGTATCCG-3′) (forward) and BY3UN (5′-CCTGTGCCCTGTTGCATTCTTC-3′) (reverse). In brief, all samples were subjected to denaturation (94 °C for 30 s), annealing (60 °C for 30 s) and extension (72 °C for 90 s) steps for 35 cycles. For the CD160 complementary DNA sequencing, the open-reading frame was amplified by PCR with BY01 and BY3UN primers and Taq High Fidelity (Life Technologies, Paisley, UK). PCR product was purified (qiaex II, Qiagen) and analyzed with the following primer sequences: forward primers BY01 (5'-TGCAGGATGCTGTTGGAACCC-3') and BY03 (3′-TCAGCCTGAACTGAACTGAGAGTGCCTTC-5′). reverse primers BY02 (5'-CAGCTGAGACTTAAAAGGGATC-3'), BY04 (3'-CACCAACACCATCTATCCCAG-5') and BY3UN (5′-CCTGTGCCCTGTTGCATTCTTC-3′).
Patients
CLL was diagnosed in 187 patients including correlation with BM aspirate, trephine, lymph node and spleen histology, flow cytometry and karyotyping when required.6, 17, 18 Patients provided informed consent. The National Research Ethics Service, East London and the City HA Local Research Ethics Committee approved non-diagnostic analyses, and written informed consent was obtained (REC reference 07/Q0604/34).
Pilot study recruitment
A pilot cohort consisted of 52 patients diagnosed with CLL. Samples were taken for routine diagnosis, then where possible at 3, 6, 9 and 12 months following treatment for the assessment of MRD. This consecutive set of patients received a variety of therapies: chlorambucil-based therapy (n=7), fludarabine (n=6), fludarabine+cyclophosphamide (n=22), fludarabine+cyclophosphamide+rituximab (n=11), alemtuzumab (n=6). Following completion of therapy, response assessment using the MRD flow assay was compared against qualitative RT-PCR.
MRD assessment by CD160FCA in paired BM and PB (Pilot Cohort)
In a subset of 35 patients from the pilot cohort, assessments of MRD using CD160FCA were performed in paired blood and BM samples prior, during or within 3 months after treatment. Sample preparation was identical for both PB and BM samples and run using the described sequential gating strategy.
Prospective validation cohort
One hundred and thirty-five patients were followed prospectively to validate the pilot data: a subset (n=38) underwent direct comparison against the current gold standard ISA approach; and a separate group of patients were assessed for outcomes (n=97). In the CD160FCA comparison with the European Research Initiative on CLL ISA methodology, a total of 48 specimens (from 38 patients), were simultaneously analyzed in four different centers. Two centers (center 1 (C1) and C2) used the ISA described by Rawstron et al,10 according to their standard operating procedures, whereas two other centers (C3 and C4) used the CD160FCA.13 Patients were recruited irrespective of their treatment regimen: fludarabine+cyclophosphamide (n=11), fludarabine+cyclophosphamide+rituximab (n=9), fludarabine+cyclophosphamide+rituximab-Lumiliximab (n=3), Chlorambucil-R (n=6), Chlorambucil-Ofatumuab (n=2), Alemtuzumab (n=3), allogeneic hemopoietic stem cell transplant (HSCT)(n=4).
Complete outcome data were available on 97/135 patients, who were repeatedly assessed as part of their ongoing monitoring for MRD by CD160FCA. Patients were assessed for time to disease detection (TTD) by CD160FCA, and correlated with known prognostic indicators (ZAP-70, CD38, cytogenetics and serum beta-2-microglobulin, β2M), as well as event-free survival (EFS, defined as time to next treatment, asymptomatic relapse or death). EFS and MRD kinetics for this cohort of patients were evaluated by Kaplan–Meier estimates and log-rank tests for trend based on first-line treatment (patient characteristics in Table 1).
Statistical analysis
Standard epidemiological approaches were used to calculate diagnostic indices of sensitivity and specificity. To determine non-random associations, the two-tailed nonparametric Fisher’s exact test was used. Non-parametric correlation coefficient r (Spearman Rank) was calculated to compare the MRD results obtained from the different centers. To determine differences in MFI of MRD populations, the two-tailed t-test based on the mean±s.e.m. was used. Comparison of Kaplan–Meier survival curves determining EFS and TTD was obtained by the nonparametric Mantel-Cox log-rank test. Levels of significance were determined as <0.05. The hazard ratio (HR) was derived from the Cox regression model indicating any hazard or chance of adverse or beneficial events.19 Statistical analysis was performed using GraphPad PRISM 5.0d for Macintosh (GraphPad software, CA, USA) and SPSS Statistics for Macintosh, Version 22.0 (Armonk, NY: IBM Corp).
Results
Simulation of MRD
The feasibility of using CD160FCA to detect MRD for CLL was assessed with a simulation assay. Samples from five patients with CLL were serially diluted in normal leukocytes, such that the CLL cells represented an actual tumor burden as low as 0.01% of the total lymphocyte population. A lower threshold of 10−4 represents the cutoff below which patients are known to have a superior outcome,20 therefore the detection of MRD to levels <0.01% were recorded as <10−4. Qualitative PCR for CD160 transcripts was also performed on purified B-cells (Supplementary Figure 1). Normalizing the data to the true malignant expression being 100%, results demonstrated a good correlation between theoretical (calculated from 100%) and observed MRD (actual analyzed MRD) (Figure 1a; R2=0.98). Light-chain (LC) restriction was detectable by conventional methodologies within the CD5+CD19+ B-cell population, but only when the CLL tumor burden represented ~40% of B-cells (data not shown). This demonstrates the limited utility of LC-restriction when dealing with low B-cell numbers. CLL cells could be reliably identified using CD160FCA to a level of 10−4 (Figure 1a). However, the CD160FCA was able to detect MRD to a level of 10−5 (Supplementary Figure 2).
MRD detection by CD160FCA and correlation with PCR in the pilot cohort. (a) The theoretical MRD expression (calculated, x axis) is plotted against the observed MRD expression (actual analyzed MRD analysis, y axis) by CD160FCA. The results are the percentage expression of CD160 of the total leukocyte population. The comparative RT-PCR results are demonstrated at the top of the graph above the corresponding dilution. The R2 linear regression value for observed expression vs theoretical expression was R2=0.98. (b) Separation of MRD-positive and MRD-negative populations based on CD160FCA assessment (P=0.01). (c) MRD assessment in paired peripheral blood and bone marrow CLL cells as determined by the CD160FCA (n=47), irrespective of the treatment regimen (Spearman Rank R=0.87, P<0.0001).
Pilot cohort—MRD assessment by CD160FCA and RT-PCR for CD160
A pilot cohort was recruited consisting of 52 cases with MRD analysis performed after a minimum of 3 months following cessation of therapy. Using 10−4 as the cutoff, CD160FCA identified two groups of MRD-positive or -negative patients (P=0.01, Figure 1b). RT-PCR for CD160 was performed on purified CD19+ B-cells (Supplementary Figure 1), which demonstrated concordant results with CD160FCA in 94% of patients. There was a strong statistical correlation between the CD160FCA and PCR results (Fisher's exact test P<0.0001, odds ratio 282).
BM or PB for MRD detection
Within a subset of the pilot cohort, MRD levels were assessed in paired PB and BM samples from 35 patients utilizing CD160FCA where results were within the quantitative range. Concordant PB and BM results were demonstrated in 30/35 cases, irrespective of the treatment regimen (P<0.001). A significant correlation was observed between PB and BM CD160FCA results (Spearman rank correlation R=0.87, P<0.001) (Figure 1c).
Prospective validation of CD160FCA
A total of 135 patients were prospectively evaluated for MRD by the CD160FCA. The separation of MRD-negative and -positive cases by CD160FCA is shown in Figure 1b, which validates the results observed in the pilot cohort.
CD160FCA vs ISA methodology in the validation cohort
A direct comparison between CD160FCA and ISA was performed involving four centers, using 49 samples from 38 patients. Results were calculated locally and then reported centrally. Operators produced concordant results in 44/49 samples (89.7%), with 3/49 cases being CD160FCApos but ISAneg, and 2/49 cases being CD160FCAneg but ISApos. MRD results were reported either as a percentage or the absolute number of cells. Regardless of the reporting method, there was a strong correlation between the two methodologies (Spearman r=0.91, P<0.001, Figure 2a). Assessing assay variability between centers performing the same methodology, the CD160FCA centers had a higher level of concordance vs the ISA methodology (Spearman r=0.94 and 0.90, respectively, P<0.01).
MRD detection by CD160FCA vs International Standardized Protocols in the validation cohort. (a) Comparative assessment of CD160FCA MRD positivity as a percentage of total leucocytes against the ISA methodology (n=48 samples, P<0.001, R=0.91). (b) MRD-positive and -negative cohorts based on the median fluorescence intensity (MFI) of the CD2neg/CD5+/CD19+/CD23+/CD160+ population.
A potential advance in the analysis and reporting of MRD data would be the use of MFI expression instead of percentage positivity. The intensity of CD160 expression was assessed for both centers performing the CD160FCA and each had significantly different MFIs between their MRD-positive and -negative patients (P=0.0002 and P=0.0005, Figure 2b). When combining the results from both centers, the mean (±s.e.m.) for MRD-positive patients was 341.3 (±52.61) and 76.38 (±12.14) if MRD was negative. Despite the differences in MFI between both centers, attributable to the use of different analysers and voltage setup, the MFI between MRD-negative and -positive populations was still significantly different.
MRD detection by CD160FCA correlates with known prognostic markers in CLL
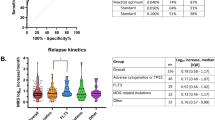
TTD following therapy, as detected by the CD160FCA in patients in CR or PR, was evaluated by univariate analysis for known prognostic markers in 97 patients, namely, cytogenetics, CD38 expression, ZAP-70 expression and serum β2M levels.21, 22 Univariate analysis demonstrated that poor risk cytogenetic status: 17p deletion (n=23) or 11q deletion (n=11) had a significantly shorter TTD (2.95 and 4.01 months, respectively) when compared with trisomy 12 (8.02 months; n=6), normal karyotype (20.97 months n=17) and 13q deletion (25.02 months, log-rank P<0.01, Figure 3a, n=40).
Correlation of time to detection of CLL by CD160FCA with known indicators of prognostication. (a) Time to detection (TTD) based on cytogenetic abnormality at diagnosis (n= 142). Median TTD: 13qdel was 25 months (n=40); normal karyotype was 21 months (n=17); Trisomy 12 was 8 months (n=6); 11qdel was 4 months (n=11); and p53 mutation/17pdel was 3 months (n=23). (b) TTD based on CD38 expression at diagnosis was shorter for CD38-positive patients (n=44) than those CD38-negative (n=53) (6 months vs 22 months, respectively, HR: 1.38, P=0.04). (c) TTD based on beta-2-microglobulin (β2M, n=89). Median TTD for elevated β2M: 6 months (n=68) vs normal β2M: 28 months (n=21, HR 2.32, P<0.01). (d) TTD based on ZAP-70 expression. Median TTD for high expression: 9 months (n=26) vs low/absent protein expression 25 months (n=42, HR: 1.65, P=0.04).
CD38 status and elevated β2M prior to treatment were predictors of TTD based on residual disease assessment by the CD160FCA: CD38+ (n=44) vs CD38- (n=53): 6.05 vs 21.99 months; P=0.04, HR: 1.38 (Figure 3b); β2M elevated (n=68) vs β2M normal (n=21): 6.03 vs 28.01 months; P<0.01, HR: 2.32 (Figure 3c). Those patients who were ZAP-70 positive had shorter TTD compared with those who were negative: ZAP-70+ (n=26) vs ZAP-70- (n=42: 9.07 vs 24.98 months; P=0.04, HR: 1.65 (Figure 3d).
CD160FCA MRD positivity correlates with shorter EFS following first-line treatment
Within the validation cohort, complete outcome data were available on 97 patients, who were followed up with MRD assessment using CD160FCA (Table 1). Responses were classified using the current National Cancer Institute guidelines and CD160FCA MRD assessment performed at regular intervals (range 1 to 81 months). Of 43 patients in CR, those who were CD160FCA MRD+ (n=11), had a significantly shorter EFS than patients in CR and MRD negative (n=32) (16 vs 63 months, respectively, P<0.001, HR 6.17, Figure 4a). Patients who achieved a CR but were MRD positive by CD160FCA had a median EFS similar to those patients achieving a partial remission based on iwCLL response (n=48) (12 months, Figure 4a). A trend was observed in median EFS for patients in PR and MRD positive (n=38) vs MRD negative (with PR primarily due to residual lymphadenopathy±residual cytopenias) (n=10) (12.5 months vs 25.5 months, respectively, P=0.12, HR 1.78 Supplementary Figure 3). There were six patients who did not respond to therapy. Patients who were in CR and MRD negative by the CD160FCA had a median time of 36 months to conversion to MRD positivity (Figure 4b) and a median time to next therapy of 60 months. Those patients in CR and MRD positive by CD160FCA analysis had a median time to next therapy of 15 months (P<0.001).
Kaplan–Meier analysis for event-free survival following first-line treatment. (a) Kaplan–Meier analysis of event-free survival (EFS) in patients with CLL (n=97) following first-line chemotherapy achieving a complete remission (CR)—MRD positive (yellow line) or CR-MRD negative (blue line)—or a partial remission (PR, gray dotted line). A significant difference in EFS was found between CR CD160FCA- (63 months) and CR CD160FCA+ (16 months, P<0.01) patients. Those patients achieving a PR had a median EFS of 12 months. (b) Kaplan–Meier plot demonstrating conversion for those patients in CR and MRD negative to detectable disease, that is, CD160FCA MRD positive. The median time to convert to MRD-positive status was 36 months (95%CI demonstrated by dotted lines).
CD160FCA MRD status is an independent prognostic marker of EFS following first-line treatment
In univariate analysis (n=97, except ZAP-70 where n=69), all the parameters assessed—MRD assessment by CD160FCA, cytogenetic status, Zap-70, CD38, elevated β2M and clinical response—were predictive of EFS. However, in multivariate analysis, only MRD assessment by CD160FCA (P<0.01, HR 5.0, Table 2) and Zap-70, status remained independent predictors of EFS (P=0.04, HR 1.90, Table 2). As ZAP-70 data were not available in 28 cases, Cox regression mutivariate analysis (n=97) was performed omitting ZAP-70, which demonstrated that not only MRD positivity by CD160FCA remained significant (P=0.03, HR 2.41), but also cytogenetic status (P=0.01, HR 2.04) was significant.
In terms of cytogenetic prognostication, patients with good-risk/intermediate cytogenetics (13q-, +12, normal karyotype) had a longer EFS following first-line therapy compared with poor-risk patients (17p- and 11q-; 38.5 vs 9.99 months, respectively, P<0.01). Interestingly, patients with good-risk cytogenetics could be further stratified based on their CD160FCA MRD status: MRD-positive patients (n=25) had significantly worse EFS vs MRD-negative patients (n=32) (18 vs 61 months, P<0.01, Supplementary Figure 4).
Discussion
Current chemo-immunotherapy regimens have led to a paradigm shift in CLL management with successful eradication of the disease burden, achievement of CR and prolongation of survival. However, conventional CR can be associated with a significant residual disease burden and MRD assessment—as evaluated by sensitive flow cytometric and molecular techniques—can further define responses in patients in CR to those as MRD-negative CR with significantly improved PFS and OS compared with those who achieve CR but are MRD positive.8, 20 This study assesses the impact and prognostic significance of MRD in CLL utilizing a simplified assay, in terms of both the number of surface proteins targeted—thereby reducing the number of monoclonal antibodies—and a simpler gating strategy. This was achieved by incorporating the B-lineage tumor-specific biomarker, CD160,13, 16 for MRD detection using a new immunophenotypic assay (CD160FCA).
The real-world data reported in this study derive from patients with CLL, treated and followed up at our institution, regardless of the treatment regimen. MRD testing was performed in routine diagnostic laboratories, with MRD positivity defined as a level of MRD >0.01% (10−4; that is, >1 in 10 000 cells). The use of CD160 has simplified the quantification of MRD to a level of at least 10−4 in both PB and BM (Figure 1b), with the potential to detect to a level of <0.001% (Supplementary Figure 2). Furthermore, it remains informative even following treatment regimens using Rituximab and Alemtuzumab. Evaluation of CD160FCA against the current gold standard (ISA) showed there was a high concordance between the two assays (P<0.01). The clinical significance of CD160FCA MRD monitoring was shown in patients following first-line treatment: those patients in CR and MRD-negative by CD160FCA had a prolonged EFS (63 months n=32) compared with patients in CR but CD160FCA MRD+ (16 months, n=11, P<0.01, Figure 4a). For those patients in PR according to the National Cancer Institute guidelines,6 outcome was similar to those patients in CR but MRD positive by CD160FCA (Figure 4a). Extending the analysis to include patients having second-line therapy or greater, MRD negativity was still associated with a prolonged EFS (CR-MRD negative 48 months vs CR-MRD positive 24 months, P<0.01, data not shown).
Achieving MRD negativity is associated with better clinical outcomes irrespective of treatment regimen and can be considered a valid clinical end point.20 In terms of EFS, biological and clinical prognostic indicators were added into a multivariate analysis model in order to observe the prognostic impact of MRD on survival. Detection of residual disease by CD160FCA and ZAP-70 status remained independent prognostic indicators of EFS when used in a multivariate analysis model (Table 2). These observations show that MRD-negative remissions, based on CD160FCA analysis, translate into improved response duration and that response duration was concordant with known pre-treatment indicators of prognostication.
In conclusion, targeting CD160, a B-lineage, tumor-specific marker, has led to the successful development of a simple single-tube, flow cytometric MRD assay, which differs from other approaches that do not contain tumor-specific targets and require more than a single tube. This approach avoids the need for a large number of monoclonal antibodies incorporated into multi-tube panels, with complex gating. We have used CD160FCA for both the diagnostic13 and MRD settings, which is of practical importance in an area where complexity and cost are continually growing. Automation of CD160FCA MRD analysis (using MFI) may allow a standardized assay to be developed, which would meet the requirements of regulatory authorities for MRD as a surrogate end point for clinical trials. The role of CD160 in MRD analysis has emerged as an independent prognostic marker, which needs to be repeated, in a larger multi-center trial.
References
Hansen MM, Andersen E, Christensen BE, Christiansen I, Geisler C, Kristensen D et al. CHOP versus prednisolone + chlorambucil in chronic lymphocytic leukemia (CLL): preliminary results of a randomized multicenter study. Nouv Rev Fr Hematol 1988; 30: 433–436.
Chemotherapeutic options in chronic lymphocytic leukemia: a meta-analysis of the randomized trials. CLL Trialists’ Collaborative Group. J Natl Cancer Inst 1999; 91: 861–868.
Rai KR, Peterson BL, Appelbaum FR, Kolitz J, Elias L, Shepherd L et al. Fludarabine compared with chlorambucil as primary therapy for chronic lymphocytic leukemia. N Engl J Med 2000; 343: 1750–1757.
Tam CS, O’Brien S, Wierda W, Kantarjian H, Wen S, Do KA et al. Long-term results of the fludarabine, cyclophosphamide, and rituximab regimen as initial therapy of chronic lymphocytic leukemia. Blood 2008; 112: 975–980.
Bosch F, Abrisqueta P, Villamor N, Terol MJ, Gonzalez-Barca E, Ferra C et al. Rituximab, fludarabine, cyclophosphamide, and mitoxantrone: a new, highly active chemoimmunotherapy regimen for chronic lymphocytic leukemia. J Clin Oncol 2009; 27: 4578–4584.
Hallek M, Cheson BD, Catovsky D, Caligaris-Cappio F, Dighiero G, Dohner H et al. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute-Working Group 1996 guidelines. Blood 2008; 111: 5446–5456.
O’Brien SM, Kantarjian HM, Thomas DA, Cortes J, Giles FJ, Wierda WG et al. Alemtuzumab as treatment for residual disease after chemotherapy in patients with chronic lymphocytic leukemia. Cancer 2003; 98: 2657–2663.
Moreton P, Kennedy B, Lucas G, Leach M, Rassam SM, Haynes A et al. Eradication of minimal residual disease in B-cell chronic lymphocytic leukemia after alemtuzumab therapy is associated with prolonged survival. J Clin Oncol 2005; 23: 2971–2979.
Bosch F, Ferrer A, Villamor N, Gonzalez M, Briones J, Gonzalez-Barca E et al. Fludarabine, cyclophosphamide, and mitoxantrone as initial therapy of chronic lymphocytic leukemia: high response rate and disease eradication. Clin Cancer Res 2008; 14: 155–161.
Rawstron AC, Villamor N, Ritgen M, Bottcher S, Ghia P, Zehnder JL et al. International standardized approach for flow cytometric residual disease monitoring in chronic lymphocytic leukaemia. Leukemia 2007; 21: 956–964.
Rawstron AC, Bottcher S, Letestu R, Villamor N, Fazi C, Kartsios H et al. Improving efficiency and sensitivity: European Research Initiative in CLL (ERIC) update on the international harmonised approach for flow cytometric residual disease monitoring in CLL. Leukemia 2013; 27: 142–149.
van Dongen JJ, Langerak AW, Bruggemann M, Evans PA, Hummel M, Lavender FL et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia 2003; 17: 2257–2317.
Farren TW, Giustiniani J, Liu FT, Tsitsikas DA, Macey MG, Cavenagh JD et al. Differential and tumor-specific expression of CD160 in B-cell malignancies. Blood 2011; 118: 2174–2183.
Maiza H, Leca G, Mansur IG, Schiavon V, Boumsell L, Bensussan A . A novel 80-kD cell surface structure identifies human circulating lymphocytes with natural killer activity. J Exp Med 1993; 178: 1121–1126.
Bensussan A, Rabian C, Schiavon V, Bengoufa D, Leca G, Boumsell L . Significant enlargement of a specific subset of CD3+CD8+ peripheral blood leukocytes mediating cytotoxic T-lymphocyte activity during human immunodeficiency virus infection. Proc Natl Acad Sci USA 1993; 90: 9427–9430.
Liu FT, Giustiniani J, Farren T, Jia L, Bensussan A, Gribben JG et al. CD160 signaling mediates PI3K-dependent survival and growth signals in chronic lymphocytic leukemia. Blood 2010; 115: 3079–3088.
Bain BJ, Barnett D, Linch D, Matutes E, Reilly JT . Revised guideline on immunophenotyping in acute leukaemias and chronic lymphoproliferative disorders. Clin Lab Haematol 2002; 24: 1–13.
Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H et al WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues, 4th edn, IARC: Lyon, France, 2008.
Spruance SL, Reid JE, Grace M, Samore M . Hazard ratio in clinical trials. Antimicrob Agents Chemother 2004; 48: 2787–2792.
Bottcher S, Ritgen M, Fischer K, Stilgenbauer S, Busch RM, Fingerle-Rowson G et al. Minimal residual disease quantification is an independent predictor of progression-free and overall survival in chronic lymphocytic leukemia: a multivariate analysis from the randomized GCLLSG CLL8 trial. J Clin Oncol 2012; 30: 980–988.
Shanafelt TD, Call TG . Current approach to diagnosis and management of chronic lymphocytic leukemia. Mayo Clin Proc 2004; 79: 388–398.
Shanafelt TD, Geyer SM, Kay NE . Prognosis at diagnosis: integrating molecular biologic insights into clinical practice for patients with CLL. Blood 2004; 103: 1202–1210.
Acknowledgements
Authorship Contribution: TWF, JG and MF designed and performed experiments as well as analyzed the data; FTL, MGM and FW performed experiments. AP and AN provided services for comparison of methodologies; SGA designed and supervised the experiments. TWF and SGA provided funding for the experiments. TWF and SGA wrote the paper.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on Blood Cancer Journal website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Farren, T., Giustiniani, J., Fanous, M. et al. Minimal residual disease detection with tumor-specific CD160 correlates with event-free survival in chronic lymphocytic leukemia. Blood Cancer Journal 5, e273 (2015). https://doi.org/10.1038/bcj.2014.92
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bcj.2014.92
This article is cited by
-
A primer set for the rapid isolation of scFv fragments against cell surface antigens from immunised rats
Scientific Reports (2020)
-
A complementary role of multiparameter flow cytometry and high-throughput sequencing for minimal residual disease detection in chronic lymphocytic leukemia: an European Research Initiative on CLL study
Leukemia (2016)