Abstract
Aim:
To identify mutants of the hepatitis B virus (HBV) X (HBx) gene and investigate the effect of the natural mutant on liver cell proliferation.
Methods:
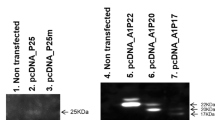
We identified natural mutants of the HBx gene from 188 sera and 48 tissues of Chinese patients infected with HBV by PCR, respectively. Based on the identification of the mutants of HBx gene, we cloned the fragments of the mutants into the pcDNA3 vector. The biological activities of the mutants were investigated.
Results:
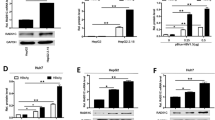
We identified a natural mutant of the HBx gene with deletion from 382 to 401 base pairs from 3 sera out of 188 patients, which resulted in the expression deletion of the HBx protein from the 128th amino acid at the COOH terminal. The similar mutant with deletion from 382 base pair at the COOH terminal was identified from 5 cases of genomes out of 48 hepatocellular carcinoma tissues. Regarding the biological activities of the mutant, we found that the mutant of the HBx protein failed to induce apoptosis by transient transfection, but promoted proliferation of human liver immortalized L-O2 cells by stable transfection, compared with the wild-type HBx protein. The data showed that the proliferation of the mutant stably-transfected L-O2-X-Sera cells and fragment stably-transfected L-O2-XΔ127 cells was enhanced by the BrdU incorporation assay and flow cytometry analysis. Lu-ciferase reporter gene assay showed that the transcriptional activities of NF-κB, survivin, and human telomerase reverse transcriptase were upregulated, and Western blot analysis revealed that the expression levels of c-Myc and proliferating cell nuclear antigen (PCNA) were upregulated in the cells.
Conclusion:
Our findings suggest that the natural HBx mutant truncated 27 amino acids at the COOH terminal promotes cell proliferation.
Similar content being viewed by others
Article PDF
References
Zhang X, Zhang W, Ye L . Pathogenesis of hepatitis B virus infection. Future Virol 2006; 1: 637–47.
Zhang X, Zhang H, Ye L . Effects of hepatitis B virus X protein on the development of Liver cancer. J Lab Clin Med 2006; 147: 58–66.
Tanaka Y, Kanai F, Kawakami T, Tateishi K, Ijichi H, Kawabe T, et al. Interaction of the hepatitis B virus X protein (HBx) with heat shock protein 60 enhances HBx-mediated apoptosis. Biochem Biophys Res Commun 2004; 318: 461–9.
León B, Taylor L, Vargas M, Luftig RB, Albertazzi F, Herrero L, et al. HBx M130K and V131I (T-A) mutations in HBV genotype F during a follow-up study in chronic carriers. Virol J 2005; 2: 60–9.
Chen GG, Li MY, Ho RL, Chak EC, Lau WY, Lai PB . Identification of hepatitis B virus X gene mutation in Hong Kong patients with hepatocellular carcinoma. J Clin Virol 2005; 34: 7–12.
Tu H, Bonura C, Giannini C, Mouly H, Soussan P, Kew M. et al. Biological impact of natural COOH-terminal deletions of hepatitis B virus X protein in hepatocellular carcinoma tissues. Cancer Res 2001; 61: 7803–10.
Zhang H, Wu L, Liu S, Shao X, Yang Z, Zhang X, et al. Examination of anti-HBx in sera from patients of chronic hepatitis B, liver cirrhosis and hepatocellular carcinoma and its clinical significance. Chin J Lab Med 2007; 30: 292–6.
Minami M, Poussin K, Brechot C, Paterlini P . A novel PCR technique using Alu-specific primers to identify unknown flanking sequences from the human genome. Genomics 1995; 29: 403–8.
Sirma H, Giannini C, Poussin K, Paterlini P, Kremsdorf D, Bréchot C . Hepatitis B virus X mutants, present in hepatocellular carcinoma tissue a brogate both the antiprolifera tive and transactivation effects of HBx. Oncogene 1999; 18: 4848–59.
Park SG, Lee SM, Jung G . Antisense oligodeoxynucleotides targeted against molecular chaperonin Hsp60 block human hepatitis B virus replication. J Biol Chem 2003; 278: 39 851–7.
Minami M, Poussin K, Brechot C, Paterlini P . A novel PCR technique using Alu-specific primers to identify unknown flanking sequences from the human genome. Genomics 1995; 29: 403–8.
Murakami Y, Minami M, Daimon Y, Okanoue T . Hepatitis B virus DNA in liver, serum, and peripheral blood mononuclear cells after the clearance of serum hepatitis B virus surface antigen. J Med Virol 2004; 72: 203–14.
Don RH, Cox PT, Wainwright BJ, Baker K, Mattick JS . ‘Touchdown’ PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res 1991; 19: 4008.
Wang F, Sha L, Zhang W, Wu L, Qiao L, Li N, et al. Involvement of hepatitis B X-interacting protein (HBXIP) in proliferation regulation of cells. Acta Pharmacol Sin 2007; 28: 431–8.
Hwang GY, Lin CY, Huang LM, Wang YH, Wang JC, Hsu CT, et al. Detection of the hepatitis B virus X protein (HBx) antigen and anti-HBx antibodies in cases of human hepatocellular carcinoma. J Clin Microbiol 2003; 41: 5598–603.
Tang NH, Chen YL, Wang XQ, Li XJ, Yin FZ, Wang XZ . Construction of IL-2 gene-modified human hepatocyte and its cultivation with microcarrier. World J Gastroenterol 2003; 9: 79–83.
Huang R, Wu T, Xu L, Liu A, Ji Y, Hu G . Upstream binding factor up-regulated in hepatocellular carcinoma is related to the survival and cisplatin-sensitivity of cancer cells. FASEB J 2002; 16: 293–301.
Liu XH, Lin J, Cao XZ, Zheng JM, Chen Y, Zhu MH . Biological impact of the COOH-terminal 40 amino acid deletions of hepatitis B virus X protein in hepatocellular carcinoma cells. Zhonghua Yi Xue Za Zhi 2005; 85: 825–30.
Tiollais P, Pourcel C, Dejean A . The hepatitis B virus. Nature 1985; 317: 489–95.
Galibert F, Mandart E, Fitoussi F, Tiollais P, Charnay P . Nucleotide sequence of the hepatitis B virus genome (subtype ayw) cloned in E coli. Nature 1979; 281: 646–50.
Xu R, Zhang X, Zhang W, Fang Y, Zheng S, Yu X . Association of human APOBEC3 cytidine deaminases with the generation of hepatitis virus B x antigen mutants and hepatocellular carcinoma. Hepatol 2007; 46: 1810–20.
Arii M, Takada S, Koike K . Identification of three essential regions of hepatitis B virus X protein for trans-activation function. Oncogene 1992; 7: 397–403.
Seeger C, Mason WS . Hepatitis B virus biology. Microbiol Mol Biol Rev 2000; 64: 51–68.
Chen GG, Li MY, Ho RL, Chak EC, Lau WY, Lai PB . Identification of hepatitis B virus X gene mutation in Hong Kong patients with hepatocellular carcinoma. J Clin Virol 2005; 34: 7–12.
Yeo W, Zhong S, Chan PK, Ho WM, Wong HT, Chan AS, et al. Sequence variations of precore/core and precore promoter regions of hepatitis B virus in patients with or without viral reactivation during cytotoxic chemotherapy. J Viral Hepat 2000; 7: 448–58.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Project supported in part by grants from the National Basic Research Program of China (973 Program, No 2007CB914800) and the National Natural Science Foundation of China (No 30670959).
Rights and permissions
About this article
Cite this article
Zhang, H., Shan, Cl., Li, N. et al. Identification of a natural mutant of HBV X protein truncated 27 amino acids at the COOH terminal and its effect on liver cell proliferation. Acta Pharmacol Sin 29, 473–480 (2008). https://doi.org/10.1111/j.1745-7254.2008.00764.x
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1111/j.1745-7254.2008.00764.x
Keywords
This article is cited by
-
Complex lithology prediction using mean impact value, particle swarm optimization, and probabilistic neural network techniques
Acta Geophysica (2020)
-
Hepatitis B virus X gene mutants emerge during antiviral therapy and increase cccDNA levels to compensate for replication suppression
Hepatology International (2020)
-
The Fragment HMGA2-sh-3p20 from HMGA2 mRNA 3′UTR Promotes the Growth of Hepatoma Cells by Upregulating HMGA2
Scientific Reports (2017)
-
USP16 Downregulation by Carboxyl-terminal Truncated HBx Promotes the Growth of Hepatocellular Carcinoma Cells
Scientific Reports (2016)
-
Variation of molybdenum isotopes in molybdenite from porphyry and vein Mo deposits in the Gangdese metallogenic belt, Tibetan plateau and its implications
Mineralium Deposita (2016)