Abstract
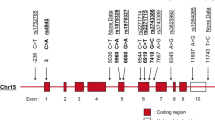
Several lines of evidence suggest interleukin-10 gene (IL-10) is a candidate gene in susceptibility to systemic lupus erythematosus (SLE). We investigated the association of IL-10 promoter single-nucleotide polymorphisms (SNPs) (−3575T/A, −2849G/A, −2763C/A, −1082A/G, −819T/C and −592A/C) and microsatellites (IL10.R, IL10.G) with SLE in 554 Hong Kong Chinese patients and 708 ethnically matched controls. Six haplotypes (hts) were identified from the SNPs. The genotype distribution of the ht1 (T-C-A-T-A), which is associated with low IL-10 production, was different in patients and controls (P=0.009). The homozygous genotype of non-ht1 was significantly increased in patients (P=0.009, odds ratio (OR)=1.80, 95% CI: 1.15–2.82). The frequency of IL10.G4 of IL10.G was also significantly increased in patients (P=0.017, OR=2.53, 95% CI: 1.18–5.40). We found that the homozygous non-ht1 combined with short allele (CA repeat number ⩽21) of IL10.G has a dose-dependent effect on SLE susceptibility: non-ht1/non-ht1 with homozygous short allele showed a higher OR (OR=4.11, 95% CI: 1.27–13.2, P=0.018) of association with SLE than the genotype of non-ht1/non-ht1 with heterozygous short/long allele (OR=2.98, 95% CI: 1.26–7.07, P=0.013) and homozygous long allele (OR=1.05, 95% CI: 0.62–1.78, P=0.848). The frequency of non-ht1 was significantly increased in patients with serositis (P<0.0001, OR=2.42, 95% CI: 1.55–3.80). In conclusion, the high expression promoter genotype is associated with SLE in Chinese.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 digital issues and online access to articles
$119.00 per year
only $19.83 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Shlomchik MJ, Craft JE, Mamula MJ . From T to B and back again: positive feedback in systemic autoimmune disease. Nat Rev Immunol 2001; 1: 147–153.
Deapen D, Escalante A, Weinrib L et al. A revised estimate of twin concordance in systemic lupus erythematosus. Arthritis Rheum 1992; 35: 311–318.
Lawrence JS, Martins CL, Drake GL . A family survey of lupus erythematosus. 1. Heritability. J Rheumatol 1987; 14: 867–869.
Vyse TJ, Kotzin BL . Genetic basis of systemic lupus erythematosus. Curr Opin Immunol 1996; 8: 843–851.
Morel L, Rudofsky UH, Longmate JA, Schiffenbauer J, Wakeland EK . Polygenic control of susceptibility to murine systemic lupus erythematosus. Immunity 1994; 1: 219–229.
Wakeland EK, Wandstrat AE, Liu K, Morel L . Genetic dissection of systemic lupus erythematosus. Curr Opin Immunol 1999; 11: 701–707.
Kelly JA, Moser KL, Harley JB . The genetics of systemic lupus erythematosus: putting the pieces together. Genes Immun 2002; 3 (Suppl 1): S71–S85.
Hawkins BR, Wong KL, Wong RWS, Chan KH, Dunckley H, Serjeantson SW . Strong association between the major histocompatibility complex and systemic lupus erythematosus in Southern Chinese. J Rheumatol 1987; 14: 1128–1131.
Lau YL, Lau CS, Chan SY, Karlberg J, Turner MW . Mannose-binding protein in Chinese patients with systemic lupus erythematosus. Arthritis Rheum 1996; 39: 706–708.
Ip WK, Chan SY, Lau CS, Lau YL . Association of systemic lupus erythematosus with promoter polymorphisms of the mannose-binding lectin gene. Arthritis Rheum 1998; 41: 1663–1668.
Ratnoff WD . Inherited deficiencies of complement in rheumatic diseases. Rheum Dis Clin N Am 1996; 22: 75–94.
Duits AJ, Bootsma H, Derksen RH et al. Skewed distribution of IgG Fc receptor IIa (CD32) polymorphism is associated with renal disease in systemic lupus erythematosus patients. Arthritis Rheum 1995; 39: 1832–1836.
Wilson AG, Gordon C, diGiovine FS et al. A genetic association between systemic lupus erythematosus and tumor necrosis factor alpha. Eur J Immunol 1994; 24: 191–195.
Prokunina L, Castillejo-Lopez C, Oberg F et al. A regulatory polymorphism in PDCD1 is associated with susceptibility to systemic lupus erythematosus in humans. Nat Genet 2002; 32: 666–669.
Gaffney PM, Kearns GM, Shark KB et al. A genome-wide search for susceptibility genes in human systemic lupus erythematosus sib-pair families. Proc Natl Acad Sci USA 1998; 95: 14875–14879.
Gaffney PM, Ortmann WA, Selby SA et al. Genome screening in human systemic lupus erythematosus: results from a second Minnesota cohort and combined analysis of 187 sib-pair families. Am J Hum Genet 2000; 66: 547–556.
Gray-McGuire C, Moser KL, Gaffney PM et al. Genome scan of human systemic lupus erythematosus by regression modeling: evidence of linkage and epistasis at 4p16–15.2. Am J Hum Genet 2000; 67: 1460–1469.
Lindqvist AK, Steinsson K, Johanneson B et al. A susceptibility locus for human systemic lupus erythematosus (hSLE1) on chromosome 2q. J Autoimmun 2000; 14: 169–178.
Shai R, Quismorio Jr FP, LI L et al. Genome-wide screen for systemic lupus erythematosus susceptibility genes in multiplex families. Hum Mol Genet 1999; 8: 639–644.
Moser KL, Neas BR, Salmon JE et al. Genome scan of human systemic lupus erythematosus: evidence for linkage on chromosome 1q in African-American pedigrees. Proc Natl Acad Sci USA 1998; 95: 14869–14874.
Johanneson B, Lima G, von Salome J et al. A major susceptibility locus for systemic lupus erythemathosus maps to chromosome 1q31. Am J Hum Genet. 2002; 71: 1060–1071.
Lander ES, Kruglyak L . Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 1995; 11: 241–247.
Tsao BP, Cantor RM, Kalunian KC et al. Evidence for linkage of a candidate chromosome 1 region to human systemic lupus erythematosus. J Clin Invest 1997; 99: 725–731.
Fiorentino DF, Bond MW, Mosmann TR . Two types of mouse T helper cell IV. Th2 clones secrete a factor that inhibits cytokine production. J Exp Med 1989; 170: 2081–2095.
Taga K, Tosato G . IL-10 inhibits T cell proliferation and IL-2 production. J Immunol 1992; 148: 1143–1148.
Ding L, Linsley PS, Huang LY, Germain RN, Shevach EM . IL-10 inhibits macrophage costimulatory activity by selectively inhibiting the up regulation of B7 expression. J Immunol 1993; 151: 1224–1234.
Moore KW, de Waal Malefyt R, Coffman RL, O'Garra A . Interleukin-10 and the interleukin-10 receptor. Annu Rev Immunol 2001; 19: 683–765.
Llorente L, Richaud-Patin Y, Wijdenes J et al. Spontaneous production of interleukin-10 by B lymphocytes and monocytes in systemic lupus erythematosus. Eur Cytokine Network 1993; 4: 421–427.
Grondal G, Gunnarsson I, Ronnelid J, Rogberg S, Klareskog L, Lundberg I . Cytokine production, serum levels and disease activity in systemic lupus erythematosus. Clin Exp Rheumatol 2000; 18: 565–570.
Llorente L, Richaud-patin Y, Couderc J et al. Dysregulation of interleukin-10 production in relatives of patients with systemic lupus erythematosus. Arthritis Rheum 1997; 40: 1429–1435.
Ishida H, Muchamuel T, Sakaguchi S, Andrade S, Menon S, Howard M . Continuous adminstration of anti-interleukin 10 antibodies delays the onset of autoimmunity in NZB/W F1 mice. J Exp Med 1994; 179: 305–310.
Westendorp RG, Langermans JA, Huizinga TW et al. Genetic influence on cytokine production and fatal meningococcal disease. Lancet 1997; 349: 170–173.
Turner DM, Williams DM, Sankaran D, Lazarus M, Sinnott PJ, Hutchinson IV . An investigation of polymorphism in the interleukin-10 gene promoter. Eur J Immunogenet 1997; 24: 1–8.
Eskdale J, Keijsers V, Huizinga T, Gallagher G . Microsatellite alleles and single nucleotide polymorphisms (SNP) combine to form four major haplotype families at the human interleukin-10 (IL-10) locus. Genes Immun 1999; 1: 151–155.
D'Alfonso S, Rampi M, Rolando V, Giordano M, Momigliano-Richiardi P . New polymorphisms in the IL-10 promoter region. Genes Immun 2000; 1: 231–233.
Gibson AW, Edberg JC, Wu J, Westendorp RG, Huizinga TW, Kimberly RP . Novel single nucleotide polymorphisms in the distal IL-10 promoter affect IL-10 production and enhance the risk of systemic lupus erythematosus. J Immunol 2001; 166: 3915–3922.
Lazarus M, Hajeer AH, Turner D et al. Genetic variation in the interleukin 10 gene promoter and systemic lupus erythematosus. J Rheumatol 1997; 24: 2314–2317.
Kube D, Platzer C, von Knethen A et al. Isolation of the human IL 10 promoter: characterization of the promoter activity in Burkitt's lymphoma cell lines. Cytokine 1995; 7: 1–7.
Edwards-Smith CJ, Jonsson JR, Purdie DM, Bansal A, Shorthouse C, Powell EE . Interleukin-10 promoter polymorphism predicts initial response of chronic hepatitis C to interferon alfa. Hepatology 1999; 30: 526–530.
Crawley E, Kay R, Sillibourne J, Patel P, Hutchinson I, Woo P . Polymorphic haplotypes of the interleukin-10 5′ flanking region determine variable interleukin-10 transcription and are associated with particular phenotypes of juvenile rheumatoid arthritis. Arthritis Rheum 1999; 42: 1101–1108.
Eskdale J, Gallagher G . A polymorphic dinucleotide repeat in the human IL-10 promoter. Immunogenetics 1995; 42: 444–445.
Eskdale J, Kube D, Gallagher G . A second polymorphic diuncleotide repeat in the 5′ flanking region of the human IL-10 gene. Immunogenetics 1996; 45: 82–83.
Eskdale J, Gallagher G, Verweij CL, Keijsers V, Westendorp RG, Huizinga TW . Interleukin 10 secretion in relation to human IL-10 locus haplotypes. Proc Natl Acad Sci USA 1998; 95: 9465–9470.
Mok CC, Lanchbury JS, Chan DW, Lau CS . Interleukin-10 promoter polymorphisms in Southern Chinese patients with systemic lupus erythematosus. Arthritis Rheum 1998; 41: 1090–1095.
Long JC, Williams RC, Urbanek M . An E–M algorithm and testing strategy for multiple-locus haplotypes. Am J Hum Genet 1995; 56: 799–810.
D'Alfonso S, Giordano M, Mellai M et al. Association tests with systemic lupus erythematosus (SLE) of IL10 markers indicate a direct involvement of a CA repeat in the 5′ regulatory region. Genes Immun 2002; 3: 454–463.
Eskdale J, Wordsworth P, Bowman S, Field M, Gallagher G . Association between polymorphisms at the human IL-10 locus and systemic lupus erythematosus. Tissue Antigens 1997; 49: 635–639.
Mehrian R, Quismorio Jr FP, Strassmann G et al. Synergistic effect between IL-10 and bcl-2 genotypes in determining susceptibility to systemic lupus erythematosus. Arthritis Rheum 1998; 41: 596–602.
D'Alfonso S, Rampi M, Bocchio D, Colombo G, Scorza-Smeraldi R, Momigliano-Richiardi P . Systemic lupus erythematosus candidate genes in the Italian population; evidence for a significant association with interleukin-10. Arthritis Rheum 2000; 43: 120–128.
Lohmueller KE, Pearce CL, Pike M, Lander ES, Hirschhorn JN . Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nat Genet 2003; 33: 177–182.
Colhoun HM, McKeigue PM, Davey Smith G . Problems of reporting genetic associations with complex outcomes. Lancet 2003; 361: 865–872.
Temple SE, Lim E, Cheong KY et al. Alleles carried at positions −819 and −592 of the IL10 promoter affect transcription following stimulation of peripheral blood cells with Streptococcus pneumoniae. Immunogenetics 2003; 55: 629–632.
Padyukov, Hahn-Zoric M, Lau YL, Hanson LA . Different allelic frequencies of several cytokine genes in Hong Kong Chinese and Swedish Caucasians. Genes Immun 2001; 2: 280–283.
Moraes MO, Santos AR, Schonkeren JJ et al. Interleukin-10 promoter haplotypes are differently distributed in the Brazilian vs the Dutch population. Immunogenetics 2003; 54: 896–899.
Tan EM, Cohen AS, Fries JF et al. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum 1982; 25: 1271–1277.
Acknowledgements
The study was partially supported by the Edward Sai-Kim Hotung Pediatric Education and Research Fund (#38) and Outstanding Researcher Award (YLL) of The University of Hong Kong.
Author information
Authors and Affiliations
Corresponding author
Additional information
The present study was approved by the Ethics Committee of the Faculty of Medicine, The University of Hong Kong.
Rights and permissions
About this article
Cite this article
Chong, W., Ip, W., Wong, WS. et al. Association of interleukin-10 promoter polymorphisms with systemic lupus erythematosus. Genes Immun 5, 484–492 (2004). https://doi.org/10.1038/sj.gene.6364119
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.gene.6364119
Keywords
This article is cited by
-
Comprehensive assessment of the association between genes on JAK-STAT pathway (IFIH1, TYK2, IL-10) and systemic lupus erythematosus: a meta-analysis
Archives of Dermatological Research (2018)
-
Association between cytokine gene polymorphisms and tuberculosis in a Chinese population in Shanghai: a case–control study
BMC Immunology (2015)
-
Lack of association between interleukin-10, transforming growth factor-beta gene polymorphisms and juvenile-onset systemic lupus erythematosus
Clinical Rheumatology (2015)
-
Association of interleukin-10 promoter haplotypes with disease susceptibility and IL-10 levels in Mexican patients with systemic lupus erythematosus
Clinical and Experimental Medicine (2015)
-
Chance, genetics, and the heterogeneity of disease and pathogenesis in systemic lupus erythematosus
Seminars in Immunopathology (2014)