Abstract
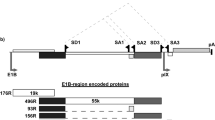
Oncolytic adenoviruses constitute a new and promising tool for cancer treatment that has been rapidly translated into clinical trials. However, minimal or absent expression of the adenovirus serotype 5 (Ad5) receptor CAR (coxsackievirus and adenovirus receptor) on cancer cells represents a major limitation for Ad5-based oncolysis. Here, we report on the resistance of CAR-negative primary melanoma cells to cell killing by wild-type Ad5 (Ad5wt) even after high titer infection, thus underlining the need for tropism-modification of oncolytic adenoviruses. We engineered a new generation of oncolytic adenoviruses that exhibit both efficient target cell infection by swapping Ad5 fiber domains with those of Ad serotype 3, which binds to a receptor distinct from CAR, and targeted virus replication. Fiber chimerism resulted in efficient cytopathicity to primary melanoma cells, which was at least 104-fold increased relative to Ad5wt. Since viral infectivity mediated by such modified viral capsids was not cell type-specific, it was pivotal to carefully restrict adenoviral replication to target cells. Towards this end, we replaced both E1A and E4 promoters of fiber chimeric viruses by tyrosinase enhancer/promoter constructs. The resulting viruses showed melanoma-specific expression of E1A and E4 and combined efficient virus replication and cell killing in melanoma cell lines and primary melanoma cells with a remarkable specificity profile that implements strong attenuation in nonmelanoma cells, including normal fibroblasts and keratinocytes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Alemany R, Balague C, Curiel DT . Replicative adenoviruses for cancer therapy. Nat Biotechnol 2000; 18: 723–727.
Zhang WW . Development and application of adenoviral vectors for gene therapy of cancer. Cancer Gene Ther 1999; 6: 113–138.
Bernt K et al. A new type of adenovirus vector that utilizes homologous recombination to achieve tumor-specific replication. J Virol 2002; 76: 10994–11002.
Miller CR et al. Differential susceptibility of primary and established human glioma cells to adenovirus infection: targeting via the epidermal growth factor receptor achieves fiber receptor-independent gene transfer. Cancer Res 1998; 58: 5738–5748.
Hemmi S et al. The presence of human coxsackievirus and adenovirus receptor is associated with efficient adenovirus-mediated transgene expression in human melanoma cell cultures. Hum Gene Ther 1998; 9: 2363–2373.
Nettelbeck DM et al. Retargeting of adenoviral infection to melanoma: combining genetic ablation of native tropism with a recombinant bispecific single-chain diabody (scDb) adapter that binds to fiber knob and HMWMAA. Int J Cancer 2004; 108: 136–145.
Reid T, Warren R, Kirn D . Intravascular adenoviral agents in cancer patients: lessons from clinical trials. Cancer Gene Ther 2002; 9: 979–986.
Barnett BG, Crews CJ, Douglas JT . Targeted adenoviral vectors. Biochim Biophys Acta 2002; 1575: 1–14.
Krasnykh VN, Douglas JT, van Beusechem VW . Genetic targeting of adenoviral vectors. Mol Ther 2000; 1: 391–405.
Nettelbeck DM et al. Novel oncolytic adenoviruses targeted to melanoma: specific viral replication and cytolysis by expression of E1A mutants from the tyrosinase enhancer/promoter. Cancer Res 2002; 62: 4663–4670.
Volk AL et al. Enhanced adenovirus infection of melanoma cells by fiber-modification: incorporation of RGD peptide or Ad5/3 chimerism. Cancer Biol Ther 2003; 2: 511–515.
Yamamoto M et al. Infectivity enhanced, cyclooxygenase-2 promoter-based conditionally replicative adenovirus for pancreatic cancer. Gastroenterology 2003; 125: 1203–1218.
Nettelbeck DM, Jerome V, Müller R . A dual specificity promoter system combining cell cycle-regulated and tissue-specific transcriptional control. Gene Therapy 1999; 6: 1276–1281.
Vigne E et al. Genetic manipulations of adenovirus type 5 fiber resulting in liver tropism attenuation. Gene Therapy 2003; 10: 153–162.
Bernt KM et al. The effect of sequestration by nontarget tissues on anti-tumor efficacy of systemically applied, conditionally replicating adenovirus vectors. Mol Ther 2003; 8: 746–755.
Gaggar A, Shayakhmetov DM, Lieber A . CD46 is a cellular receptor for group B adenoviruses. Nat Med 2003; 9: 1408–1412; Epub 2003 Oct 1419.
Kawakami Y et al. Substitution of the adenovirus serotype 5 knob with a serotype 3 knob enhances multiple steps in virus replication. Cancer Res 2003; 63: 1262–1269.
Suzuki K et al. A conditionally replicative adenovirus with enhanced infectivity shows improved oncolytic potency. Clin Cancer Res 2001; 7: 120–126.
Kanerva A et al. Enhanced therapeutic efficacy for ovarian cancer with a serotype 3 receptor-targeted oncolytic adenovirus. Mol Ther 2003; 8: 449–458.
Reynolds PN et al. Combined transductional and transcriptional targeting improves the specificity of transgene expression in vivo. Nat Biotechnol 2001; 19: 838–842.
Barnett BG, Tillman BW, Curiel DT, Douglas JT . Dual targeting of adenoviral vectors at the levels of transduction and transcription enhances the specificity of gene expression in cancer cells. Mol Ther 2002; 6: 377–385.
Kanerva A et al. A cyclooxygenase-2 promoter-based conditionally replicating adenovirus with enhanced infectivity for treatment of ovarian adenocarcinoma. Gene Therapy 2004; 11: 552–559.
Banerjee NS et al. Analyses of melanoma-targeted oncolytic adenoviruses with tyrosinase enhancer/promoter-driven E1A, E4, or both in submerged cells and organotypic cultures. Mol Cancer Ther 2004; 3: 437–449.
Fueyo J et al. A mutant oncolytic adenovirus targeting the Rb pathway produces anti-glioma effect in vivo. Oncogene 2000; 19: 2–12.
Heise C et al. An adenovirus E1A mutant that demonstrates potent and selective systemic anti-tumoral efficacy. Nat Med 2000; 6: 1134–1139.
Piepkorn M . Melanoma genetics: an update with focus on the CDKN2A(p16)/ARF tumor suppressors. J Am Acad Dermatol 2000; 42: 705–722; quiz 723–726.
Shayakhmetov DM, Lieber A . Dependence of adenovirus infectivity on length of the fiber shaft domain. J Virol 2000; 74: 10274–10286.
Segerman A et al. Adenovirus type 11 uses CD46 as a cellular receptor. J Virol 2003; 77: 9183–9191.
Sirena D et al. The human membrane cofactor CD46 is a receptor for species B adenovirus serotype 3. J Virol 2004; 78: 4454–4462.
Short JJ et al. Adenovirus serotype 3 utilizes CD80 (B7.1) and CD86 (B7.2) as cellular attachment receptors. Virology 2004; 322: 349–359.
Wu H et al. Double modification of adenovirus fiber with RGD and polylysine motifs improves coxsackievirus-adenovirus receptor-independent gene transfer efficiency. Hum Gene Ther 2002; 13: 1647–1653.
Krasnykh VN, Mikheeva GV, Douglas JT, Curiel DT . Generation of recombinant adenovirus vectors with modified fibers for altering viral tropism. J Virol 1996; 70: 6839–6846.
Bergelson JM et al. Isolation of a common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science 1997; 275: 1320–1323.
Rivera AA et al. Mode of transgene expression after fusion to early or late viral genes of a conditionally replicating adenovirus via an optimized internal ribosome entry site in vitro and in vivo. Virology 2004; 320: 121–134.
Acknowledgements
This work was supported by the Deutsche Forschungsgemeinschaft (grant NE832/1 to DMN, and Graduiertenkolleg 592 to SS), the Deutsche Krebshilfe (grant 10-2186-Ne 1 to DMN) and the National Cancer Institute (grants R01 CA83821, P50 CA83591, R01 CA93796, and R01 CA94084). We are grateful to Dr D Dieckmann, Dr IJ Fidler, Dr F Noya, Dr J Price, L Rivera, Dr J Schlom, and Dr T Strong for cell lines and primary cells, to Dr J Chrobozcek for providing plasmid pBR.Ad3Fib, Dr Ruben Hernandez-Alcoceba for plasmid pUC19-E4P-, Dr Gary Ketner for the E4 antibody, and to T Uil for plasmid pNEB.PK.SnaBI.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Rivera, A., Davydova, J., Schierer, S. et al. Combining high selectivity of replication with fiber chimerism for effective adenoviral oncolysis of CAR-negative melanoma cells. Gene Ther 11, 1694–1702 (2004). https://doi.org/10.1038/sj.gt.3302346
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.gt.3302346
Keywords
This article is cited by
-
Combing oncolytic adenovirus expressing Beclin-1 with chemotherapy agent doxorubicin synergistically enhances cytotoxicity in human CML cells in vitro
Acta Pharmacologica Sinica (2018)
-
Application of conditionally replicating adenoviruses in tumor early diagnosis technology, gene-radiation therapy and chemotherapy
Applied Microbiology and Biotechnology (2016)
-
Systemically administered liposome-encapsulated Ad-PEDF potentiates the anti-cancer effects in mouse lung metastasis melanoma
Journal of Translational Medicine (2013)
-
Therapeutic Improvement of a Stroma-Targeted CRAd by Incorporating Motives Responsive to the Melanoma Microenvironment
Journal of Investigative Dermatology (2013)
-
Oncolytic adenovirus retargeted to Delta-EGFR induces selective antiglioma activity
Cancer Gene Therapy (2009)