Abstract
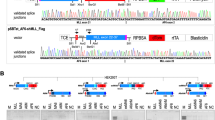
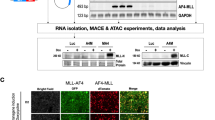
Chromosomal rearrangements of the human MLL gene are a hallmark for aggressive (high-risk) pediatric, adult and therapy-associated acute leukemias. These patients need to be identified in order to subject these patients to appropriate therapy regimen. A recently developed long-distance inverse PCR method was applied to genomic DNA isolated from individual acute leukemia patients in order to identify chromosomal rearrangements of the human MLL gene. We present data of the molecular characterization of 414 samples obtained from 272 pediatric and 142 adult leukemia patients. The precise localization of genomic breakpoints within the MLL gene and the involved translocation partner genes (TPGs) was determined and several new TPGs were identified. The combined data of our study and published data revealed a total of 87 different MLL rearrangements of which 51 TPGs are now characterized at the molecular level. Interestingly, the four most frequently found TPGs (AF4, AF9, ENL and AF10) encode nuclear proteins that are part of a protein network involved in histone H3K79 methylation. Thus, translocations of the MLL gene, by itself coding for a histone H3K4 methyltransferase, are presumably not randomly chosen, rather functionally selected.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Pui CH, Gaynon PS, Boyett JM, Chessells JM, Baruchel A, Kamps W et al. Outcome of treatment in childhood acute lymphoblastic leukaemia with rearrangements of the 11q23 chromosomal region. Lancet 2002; 359: 1909–1915.
Pui CH, Chessells JM, Camitta B, Baruchel A, Biondi A, Boyett JM et al. Clinical heterogeneity in childhood acute lymphoblastic leukemia with 11q23 rearrangements. Leukemia 2003; 17: 700–706.
Meyer C, Schneider B, Reichel M, Angermueller S, Strehl S, Schnittger S et al. Diagnostic tool for the identification of MLL rearrangements including unknown partner genes. Proc Natl Acad Sci USA 2005; 102: 449–454.
Reichel M, Gillert E, Angermuller S, Hensel JP, Heidel F, Lode M et al. Biased distribution of chromosomal breakpoints involving the MLL gene in infants versus children and adults with t(4;11) ALL. Oncogene 2001; 20: 2900–2907.
Katoh M, Katoh M . Identification and characterization of human BCL9L gene and mouse Bcl9l gene in silico. Int J Mol Med 2003; 12: 643–649.
Teuffel O, Betts DR, Thali M, Eberle D, Meyer C, Schneider B et al. Clonal expansion of a new MLL rearrangement in the absence of leukemia. Blood 2005; 105: 4151–4152.
Reichel M, Gillert E, Nilson I, Siegler G, Greil J, Fey GH et al. Fine structure of translocation breakpoints in leukemic blasts with chromosomal translocation t(4;11): the DNA damage-repair model of translocation. Oncogene 1998; 17: 3035–3044.
Richardson C, Jasin M . Frequent chromosomal translocations induced by DNA double-strand breaks. Nature 2000; 405: 697–700.
Schichman SA, Caligiuri MA, Gu Y, Strout MP, Canaani E, Bloomfield CD et al. ALL-1 partial duplication in acute leukemia. Proc Natl Acad Sci USA 1994; 91: 6236–6239.
Caligiuri MA, Strout MP, Schichman SA, Mrozek K, Arthur DC, Herzig GP et al. Partial tandem duplication of ALL1 as a recurrent molecular defect in acute myeloid leukemia with trisomy 11. Cancer Res 1996; 56: 1418–1425.
Schnittger S, Kinkelin U, Schoch C, Heinecke A, Haase D, Haferlach T et al. Screening for MLL tandem duplication in 387 unselected patients with AML identify a prognostically unfavorable subset of AML. Leukemia 2000; 14: 796–804.
Martin ME, Milne TA, Bloyer S, Galoian K, Shen W, Gibbs D et al. Dimerization of MLL fusion proteins immortalizes hematopoietic cells. Cancer Cell 2003; 4: 197–207.
Marschalek R, Nilson I, Lochner K, Greim R, Siegler G, Greil J et al. The structure of the human ALL-1/MLL/HRX gene. Leuk Lymphoma 1997; 27: 417–428.
Langer T, Metzler M, Reinhardt D, Viehmann S, Borkhardt A, Reichel M et al. Analysis of t(9;11) chromosomal breakpoint sequences in childhood acute leukemia: almost identical MLL breakpoints in therapy-related AML after treatment without etoposides. Genes Chrom Cancer 2003; 36: 393–401.
Metzler M, Brehm U, Langer T, Viehmann S, Borkhardt A, Stanulla M et al. Asymmetric multiplex-polymerase chain reaction – a high throughput method for detection and sequencing genomic fusion sites in t(4;11). Br J Haematol 2004; 124: 47–54.
Burmeister T, Marschalek R, Schneider B, Meyer C, Gökbuget N, Schwartz S et al. Monitoring minimal residual disease by quantification of genomic chromosomal breakpoint sequences in acute leukemias with MLL aberrations. Leukemia 2006, 19 January, [E-pub ahead of print].
Erfurth F, Hemenway CS, de Erkenez AC, Domer P . MLL fusion partners AF4 and AF9 interact at subnuclear foci. Leukemia 2004; 18: 92–102.
Srinivasan RS, Nesbit JB, Marrero L, Erfurth F, LaRussa VF, Hemenway CS . The synthetic peptide PFWT disrupts AF4–AF9 protein complexes and induces apoptosis in t(4;11) leukemia cells. Leukemia 2004; 18: 1364–1372.
Isnard P, Core N, Naquet P, Djabali M . Altered lymphoid development in mice deficient for the mAF4 proto-oncogene. Blood 2000; 96: 705–710.
Zeisig DT, Bittner CB, Zeisig BB, Garcia-Cuellar MP, Hess JL, Slany RK . The eleven–nineteen-leukemia protein ENL connects nuclear MLL fusion partners with chromatin. Oncogene 2005; 24: 5525–5532.
Okada Y, Feng Q, Lin Y, Jiang Q, Li Y, Coffield VM et al. hDOT1L links histone methylation to leukemogenesis. Cell 2005; 121: 167–178.
Jenuwein T, Allis CD . Translating the histone code. Science 2001; 293: 1074–1080.
Lachner M, Jenuwein T . The many faces of histone lysine methylation. Curr Opin Cell Biol 2002; 14: 286–298.
Zhang Y, Reinberg D . Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev 2001; 15: 2343–2360.
Bursen A, Moritz S, Gaussmann A, Dingermann T, Marschalek R . Interaction of AF4 wildtype and AF4·MLL fusion protein with SIAH proteins: indication for t(4;11) pathobiology? Oncogene 2004; 23: 6237–6249.
Acknowledgements
This work was made possible by and conducted within the framework of the International BFM Study Group. We thank Reinald Repp, Thorsten Langer, Jörn-D Beck, Markus Metzler und Thomas Leis for providing and sharing unpublished information of their ongoing study (Grant 2002.032.1 from the Wilhelm Sander Foundation) and Michael Karas for critical comments. This study is supported by Grant GEN-AU Child, GZ 200.071/3-VI/2a/2002 to OAH and Grant 2001.061.1 from the Wilhelm-Sander-Foundation to RM, TD and TK.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on the Leukemia website (http://www.nature.com/leu)
Supplementary information
Rights and permissions
About this article
Cite this article
Meyer, C., Schneider, B., Jakob, S. et al. The MLL recombinome of acute leukemias. Leukemia 20, 777–784 (2006). https://doi.org/10.1038/sj.leu.2404150
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.leu.2404150
Keywords
This article is cited by
-
The KMT2A recombinome of acute leukemias in 2023
Leukemia (2023)
-
MYB oncoproteins: emerging players and potential therapeutic targets in human cancer
Oncogenesis (2021)
-
The role of EVI1 gene quantification in AML patients with 11q23/MLL rearrangement after allogeneic hematopoietic stem cell transplantation
Bone Marrow Transplantation (2021)
-
The lysine methyltransferase SMYD2 is required for normal lymphocyte development and survival of hematopoietic leukemias
Genes & Immunity (2020)
-
The MLL recombinome of acute leukemias in 2017
Leukemia (2018)