Abstract
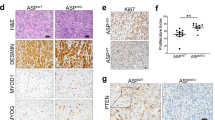
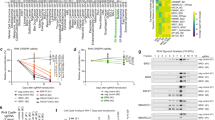
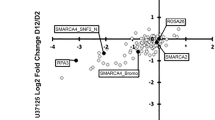
The chromosomal translocation t(2;13), characteristic for the aggressive childhood cancer alveolar rhabdomyosarcoma (aRMS), generates the chimeric transcription factor PAX3/FKHR with a well known oncogenic role. However, the molecular mechanisms mediating essential pathophysiological functions remain poorly defined. Here, we used comparative expression profiling of PAX3/FKHR silencing in vitro and PAX3/FKHR-specific gene signatures in vivo to identify physiologically important target genes. Hereby, 51 activated genes, both novel and known, were identified. We also found repression of skeletal muscle-specific genes suggesting that PAX3/FKHR blocks further differentiation of aRMS cells. Importantly, TFAP2B was validated as direct target gene mediating the anti-apoptotic function of PAX3/FKHR. Hence, we developed a pathophysiologically relevant transcriptional profile of PAX3/FKHR and identified a critical target gene for aRMS development.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Anderson J, Ramsay A, Gould S, Pritchard-Jones K . (2001). PAX3-FKHR induces morphological change and enhances cellular proliferation and invasion in rhabdomyosarcoma. Am J Pathol 159: 1089–1096.
Ayyanathan K, Fredericks WJ, Berking C, Herlyn M, Balakrishnan C, Gunther E et al. (2000). Hormone-dependent tumor regression in vivo by an inducible transcriptional repressor directed at the PAX3-FKHR oncogene. Cancer Res 60: 5803–5814.
Begum S, Emani N, Cheung A, Wilkins O, Der S, Hamel PA . (2005). Cell-type-specific regulation of distinct sets of gene targets by Pax3 and PAX3/FKHR. Oncogene 24: 1860–1872.
Bennicelli JL, Fredericks WJ, Wilson RB, Rauscher III FJ, Barr FG . (1995). Wild type PAX3 protein and the PAX3-FKHR fusion protein of alveolar rhabdomyosarcoma contain potent, structurally distinct transcriptional activation domains. Oncogene 11: 119–130.
Bernasconi M, Remppis A, Fredericks WJ, Rauscher III FJ, Schafer BW . (1996). Induction of apoptosis in rhabdomyosarcoma cells through down-regulation of PAX proteins. Proc Natl Acad Sci USA 93: 13164–13169.
Davicioni E, Finckenstein FG, Shahbazian V, Buckley JD, Triche TJ, Anderson MJ . (2006). Identification of a PAX-FKHR gene expression signature that defines molecular classes and determines the prognosis of alveolar rhabdomyosarcomas. Cancer Res 66: 6936–6946.
Edelman GM, Jones FS . (1995). Developmental control of N-CAM expression by Hox and Pax gene products. Philos Trans R Soc London B Biol Sci 349: 305–312.
Elbashir SM, Lendeckel W, Tuschl T . (2001). RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev 15: 188–200.
Epstein JA, Shapiro DN, Cheng J, Lam PY, Maas RL . (1996). Pax3 modulates expression of the c-Met receptor during limb muscle development. Proc Natl Acad Sci USA 93: 4213–4218.
Epstein JA, Song B, Lakkis M, Wang C . (1998). Tumor-specific PAX3-FKHR transcription factor, but not PAX3, activates the platelet-derived growth factor alpha receptor. Mol Cell Biol 18: 4118–4130.
Fredericks WJ, Galili N, Mukhopadhyay S, Rovera G, Bennicelli J, Barr FG et al. (1995). The PAX3-FKHR fusion protein created by the t(2;13) translocation in alveolar rhabdomyosarcomas is a more potent transcriptional activator than PAX3. Mol Cell Biol 15: 1522–1535.
Galili N, Davis RJ, Fredericks WJ, Mukhopadhyay S, Rauscher III FJ, Emanuel BS et al. (1993). Fusion of a fork head domain gene to PAX3 in the solid tumour alveolar rhabdomyosarcoma. Nat Genet 5: 230–235.
Ginsberg JP, Davis RJ, Bennicelli JL, Nauta LE, Barr FG . (1998). Up-regulation of MET but not neural cell adhesion molecule expression by the PAX3-FKHR fusion protein in alveolar rhabdomyosarcoma. Cancer Res 58: 3542–3546.
Keller C, Arenkiel BR, Coffin CM, El-Bardeesy N, DePinho RA, Capecchi MR . (2004). Alveolar rhabdomyosarcomas in conditional Pax3:Fkhr mice: cooperativity of Ink4a/ARF and Trp53 loss of function. Genes Dev 18: 2614–2626.
Khan J, Simon R, Bittner M, Chen Y, Leighton SB, Pohida T et al. (1998). Gene expression profiling of alveolar rhabdomyosarcoma with cDNA microarrays. Cancer Res 58: 5009–5013.
Lam PY, Sublett JE, Hollenbach AD, Roussel MF . (1999). The oncogenic potential of the Pax3-FKHR fusion protein requires the Pax3 homeodomain recognition helix but not the Pax3 paired-box DNA binding domain. Mol Cell Biol 19: 594–601.
Lang D, Lu MM, Huang L, Engleka KA, Zhang M, Chu EY et al. (2005). Pax3 functions at a nodal point in melanocyte stem cell differentiation. Nature 433: 884–887.
Li C, Wong WH . (2001). Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA 98: 31–36.
Margue CM, Bernasconi M, Barr FG, Schafer BW . (2000). Transcriptional modulation of the anti-apoptotic protein BCL-XL by the paired box transcription factors PAX3 and PAX3/FKHR. Oncogene 19: 2921–2929.
Mayanil CS, George D, Freilich L, Miljan EJ, Mania-Farnell B, McLone DG et al. (2001). Microarray analysis detects novel Pax3 downstream target genes. J Biol Chem 276: 49299–49309.
Moser M, Dahmen S, Kluge R, Grone H, Dahmen J, Kunz D et al. (2003). Terminal renal failure in mice lacking transcription factor AP-2 beta. Lab Invest 83: 571–578.
Moser M, Pscherer A, Roth C, Becker J, Mucher G, Zerres K et al. (1997). Enhanced apoptotic cell death of renal epithelial cells in mice lacking transcription factor AP-2beta. Genes Dev 11: 1938–1948.
Scheidler S, Fredericks WJ, Rauscher III FJ, Barr FG, Vogt PK . (1996). The hybrid PAX3-FKHR fusion protein of alveolar rhabdomyosarcoma transforms fibroblasts in culture. Proc Natl Acad Sci USA 93: 9805–9809.
Tomescu O, Xia SJ, Strezlecki D, Bennicelli JL, Ginsberg J, Pawel B et al. (2004). Inducible short-term and stable long-term cell culture systems reveal that the PAX3-FKHR fusion oncoprotein regulates CXCR4, PAX3, and PAX7 expression. Lab Invest 84: 1060–1070.
Wachtel M, Dettling M, Koscielniak E, Stegmaier S, Treuner J, Simon-Klingenstein K et al. (2004). Gene expression signatures identify rhabdomyosarcoma subtypes and detect a novel t(2;2)(q35;p23) translocation fusing PAX3 to NCOA1. Cancer Res 64: 5539–5545.
Wachtel M, Runge T, Leuschner I, Stegmaier S, Koscielniak E, Treuner J et al. (2006). Subtype and prognostic classification of rhabdomyosarcoma by immunohistochemistry. J Clin Oncol 24: 816–822.
Xia SJ, Barr FG . (2004). Analysis of the transforming and growth suppressive activities of the PAX3-FKHR oncoprotein. Oncogene 23: 6864–6871.
Zhang L, Wang C . (2006). Identification of a new class of PAX3-FKHR target promoters: a role of the Pax3 paired box DNA binding domain. Oncogene 26: 1595–1605.
Acknowledgements
We thank FG Barr (University of Pennsylvania, Philadelphia, PA) and R Fässler (Max-Planck-Institute, Münich, Germany) for providing cDNA constructs, A Patrignani (FGCZ) for excellent technical assistance with Affymetrix experiments and M Dettling for performing principal component analysis. This work was supported by Swiss National Science Foundation, grant nos. 3100–067841 and 3100–109837 and the Schweizerische Forschungsstiftung Kind und Krebs.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc).
Rights and permissions
About this article
Cite this article
Ebauer, M., Wachtel, M., Niggli, F. et al. Comparative expression profiling identifies an in vivo target gene signature with TFAP2B as a mediator of the survival function of PAX3/FKHR. Oncogene 26, 7267–7281 (2007). https://doi.org/10.1038/sj.onc.1210525
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.onc.1210525
Keywords
This article is cited by
-
Crucial role of the transcription factors family activator protein 2 in cancer: current clue and views
Journal of Translational Medicine (2023)
-
Pax3 loss of function delays tumour progression in kRAS-induced zebrafish rhabdomyosarcoma models
Scientific Reports (2022)
-
FOXF1 is required for the oncogenic properties of PAX3-FOXO1 in rhabdomyosarcoma
Oncogene (2021)
-
Co-expression of transcription factor AP-2beta (TFAP2B) and GATA3 in human mammary epithelial cells with intense, apicobasal immunoreactivity for CK8/18
Journal of Molecular Histology (2021)
-
OLIG2 is a novel immunohistochemical marker associated with the presence of PAX3/7-FOXO1 translocation in rhabdomyosarcomas
Diagnostic Pathology (2019)