Abstract
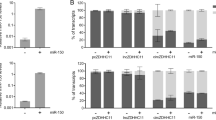
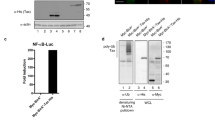
BIC is a primary microRNA (pri-miR-155) that can be processed to mature miR-155. In this study, we show the crucial involvement of protein kinase C (PKC) and nuclear factor-κB (NF-κB) in the regulation of BIC expression upon B-cell receptor triggering. Surprisingly, Northern blot analysis did not reveal any miR-155 expression upon induction of BIC expression in the Burkitt lymphoma-derived Ramos cell line, whereas other microRNAs were clearly detectable. Ectopic expression of BIC in Ramos and HEK293 cells resulted in miR-155 expression in HEK293, but not in Ramos cells, suggesting a specific block of BIC to miR-155 processing in Ramos. In line with the results obtained with Ramos, lack of miR-155 expression after induction of BIC expression was also observed in other Burkitt lymphoma cell lines, indicating a generic and specific blockade in the processing of BIC in Burkitt lymphoma. In contrast, induction of BIC expression in normal tonsillar B cells resulted in very high levels of miR-155 expression and induction of BIC expression in Hodgkin's lymphoma cell lines. It also resulted in elevated levels of miR-155. Our data provide evidence for two levels of regulation for mature miR-155 expression: one at the transcriptional level involving PKC and NF-κB, and one at the processing level. Burkitt lymphoma cells not only express low levels of BIC, but also prevent processing of BIC via an, as yet, unknown mechanism.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Calin GA, Liu CG, Sevignani C, Ferracin M, Felli N, Dumitru CD et al. (2004). MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc Natl Acad Sci USA 101: 11755–11760.
Chen CZ . (2005). MicroRNAs as oncogenes and tumor suppressors. N Engl J Med 353: 1768–1771.
Chen CZ, Li L, Lodish HF, Bartel DP . (2004). MicroRNAs modulate hematopoietic lineage differentiation. Science 303: 83–86.
Ciafre SA, Galardi S, Mangiola A, Ferracin M, Liu CG, Sabatino G et al. (2005). Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun 334: 1351–1358.
Clurman BE, Hayward WS . (1989). Multiple proto-oncogene activations in avian leukosis virus-induced lymphomas: evidence for stage-specific events. Mol Cell Biol 9: 2657–2664.
Eis PS, Tam W, Sun L, Chadburn A, Li Z, Gomez MF et al. (2005). Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci USA 102: 3627–3632.
Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N et al. (2004). The Microprocessor complex mediates the genesis of microRNAs. Nature 432: 235–240.
Haasch D, Chen YW, Reilly RM, Chiou XG, Koterski S, Smith ML et al. (2002). T cell activation induces a noncoding RNA transcript sensitive to inhibition by immunosuppressant drugs and encoded by the proto-oncogene, BIC. Cell Immunol 217: 78–86.
Hammond SM . (2006). MicroRNAs as oncogenes. Curr Opin Genet Dev 16: 4–9.
Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD . (2001). A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293: 834–838.
Hutvagner G, Zamore PD . (2002). A microRNA in a multiple-turnover RNAi enzyme complex. Science 297: 2056–2060.
Jiang J, Lee EJ, Schmittgen TD . (2006). Increased expression of microRNA-155 in Epstein-Barr virus transformed lymphoblastoid cell lines. Genes Chromosomes Cancer 45: 103–106.
Kluiver J, Haralambieva E, de Jong D, Blokzijl T, Jacobs S, Kroesen BJ et al. (2006). Lack of BIC and microRNA miR-155 expression in primary cases of Burkitt lymphoma. Genes Chromosomes Cancer 45: 147–153.
Kluiver J, Poppema S, de Jong D, Blokzijl T, Harms G, Jacobs S et al. (2005). BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas. J Pathol 207: 243–249.
Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T . (2002). Identification of tissue-specific microRNAs from mouse. Curr Biol 12: 735–739.
Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J et al. (2003). The nuclear RNase III Drosha initiates microRNA processing. Nature 425: 415–419.
Luciano DJ, Mirsky H, Vendetti NJ, Maas S . (2004). RNA editing of a miRNA precursor. RNA 10: 1174–1177.
Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U . (2004). Nuclear export of microRNA precursors. Science 303: 95–98.
Metzler M, Wilda M, Busch K, Viehmann S, Borkhardt A . (2004). High expression of precursor microRNA-155/BIC RNA in children with Burkitt lymphoma. Genes Chromosomes Cancer 39: 167–169.
Obernosterer G, Leuschner PJF, Alenius M, Marinez J . (2006). Post-transcriptional regulation of microRNA expression. RNA 12: 1161–1167.
Pfeffer S, Zavolan M, Grasser FA, Chien M, Russo JJ, Ju J et al. (2004). Identification of virus-encoded microRNAs. Science 304: 734–736.
Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE et al. (2004). A pancreatic islet-specific microRNA regulates insulin secretion. Nature 432: 226–230.
Ramkissoon SH, Mainwaring LA, Ogasawara Y, Keyvanfar K, Philip McCoy Jr J, Sloand EM et al. (2006). Hematopoietic-specific microRNA expression in human cells. Leuk Res 30: 643–647.
Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M et al. (2006). A brain-specific microRNA regulates dendritic spine development. Nature 439: 283–289.
Specht K, Richter T, Muller U, Walch A, Werner M, Hofler H . (2001). Quantitative gene expression analysis in microdissected archival formalin-fixed and paraffin-embedded tumor tissue. Am J Pathol 158: 419–429.
Suh MR, Lee Y, Kim JY, Kim SK, Moon SH, Lee JY et al. (2004). Human embryonic stem cells express a unique set of microRNAs. Dev Biol 270: 488–498.
Tam W, Hughes SH, Hayward WS, Besmer P . (2002). Avian bic, a gene isolated from a common retroviral site in avian leukosis virus-induced lymphomas that encodes a noncoding RNA, cooperates with c-myc in lymphomagenesis and erythroleukemogenesis. J Virol 76: 4275–4286.
van den Berg A, Kroesen BJ, Kooistra K, de Jong D, Briggs J, Blokzijl T et al. (2003). High expression of B-cell receptor inducible gene BIC in all subtypes of Hodgkin's lymphoma. Genes Chromosomes Cancer 37: 20–28.
Yang W, Chendrimada TP, Wang Q, Higuchi M, Seeburg PH, Shiekhattar R et al. (2006). Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat Struct Mol Biol 13: 13–21.
Zeng Y, Cullen BR . (2005). Efficient processing of primary microRNA hairpins by Drosha requires flanking nonstructured RNA sequences. J Biol Chem 280: 27595–27603.
Acknowledgements
This study was supported by a grant from the Dutch Cancer Society (RUG 01-2414)
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kluiver, J., van den Berg, A., de Jong, D. et al. Regulation of pri-microRNA BIC transcription and processing in Burkitt lymphoma. Oncogene 26, 3769–3776 (2007). https://doi.org/10.1038/sj.onc.1210147
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.onc.1210147
Keywords
This article is cited by
-
MicroRNA’s in cancer as biomarkers and therapeutic keys
ExRNA (2020)
-
Deregulation of neuronal miRNAs induced by amyloid-β or TAU pathology
Molecular Neurodegeneration (2018)
-
miR-155 in cancer drug resistance and as target for miRNA-based therapeutics
Cancer and Metastasis Reviews (2018)
-
Toxoplasma Modulates Signature Pathways of Human Epilepsy, Neurodegeneration & Cancer
Scientific Reports (2017)
-
miR expression in MYC-negative DLBCL/BL with partial trisomy 11 is similar to classical Burkitt lymphoma and different from diffuse large B–cell lymphoma
Tumor Biology (2015)