Abstract
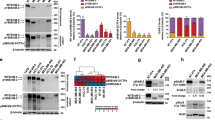
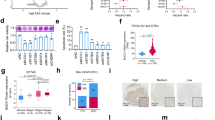
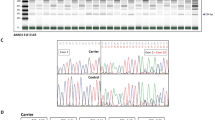
Acute myeloid leukemia (AML) 1 is often disrupted by chromosomal translocations generating oncogenic fusions in human leukemias. However, its role in epithelial cancers has not been extensively investigated. Herein, we show a marked accumulation of AML1 transcripts including a high frequency of a novel alternatively spliced AML1b transcript lacking exon 6 (AML1bDel179−242) in ovarian cancer patients. The increases in RNA transcripts for total wild-type AML1 and AML1bDel179−242 are associated with poor patient outcomes. We have shown that although both wild-type AML1b and AML1bDel179−242 are localized to nuclear speckles, AML1bDel179−242 was observed to have dramatically reduced transactivation potential with the plasminogen activator inhibitor-1 promoters and behaved as a weak dominant negative of wild-type AML1b. Wild-type AML1b was found to inhibit the growth of immortalized ovarian epithelial cells (T29) decreasing colony-forming ability. Moreover, we have identified a novel function of AML1b where it inhibits ovarian cell migration. In contrast, AML1bDel179−242 has lost the ability to inhibit both ovarian cell proliferation and migration indicating that the functional effects observed with wild-type AML1b are dependent on amino acids 179–242. Collectively, these studies suggest that deregulated alternative splicing of AML1b transcripts may potentially contribute to the pathophysiology of ovarian cancers.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
Abbreviations
- AML:
-
acute myeloid leukemia
- RHD:
-
runt homology domain
- TGFβ:
-
transforming growth factor β
- OSE:
-
ovarian epithelial cells
- PAI-1:
-
plasminogen activator inhibitor-1
References
Bae SC, Ogawa E, Maruyama M, Oka H, Satake M, Shigesada K et al. (1994). PEBP2 alpha B/mouse AML1 consists of multiple isoforms that possess differential transactivation potentials. Mol Cell Biol 14: 3242–3252.
Bartel F, Taubert H, Harris LC . (2002). Alternative and aberrant splicing of MDM2 mRNA in human cancer. Cancer Cell 2: 9–15.
Berger R . (1997). Acute lymphoblastic leukemia and chromosome 21. Cancer Genet Cytogenet 94: 8–12.
Biggs JR, Zhang Y, Peterson LF, Garcia M, Zhang DE, Kraft AS . (2005). Phosphorylation of AML1/RUNX1 regulates its degradation and nuclear matrix association. Mol Cancer Res 3: 391–401.
Dairkee SH, Ji Y, Ben Y, Moore DH, Meng Z, Jeffrey SS . (2004). A molecular ‘signature’ of primary breast cancer cultures; patterns resembling tumor tissue. BMC Genomics 5: 47.
Faleiro-Rodrigues C, Macedo-Pinto I, Pereira D, Ferreira VM, Lopes CS . (2004). Association of E-cadherin and beta-catenin immunoexpression with clinicopathologic features in primary ovarian carcinomas. Hum Pathol 35: 663–669.
Fowler M, Borazanci E, McGhee L, Pylant SW, Williams BJ, Glass J et al. (2006). RUNX1 (AML-1) and RUNX2 (AML-3) cooperate with prostate-derived Ets factor to activate transcription from the PSA upstream regulatory region. J Cell Biochem 97: 1–17.
Fujioka T, Takebayashi Y, Kihana T, Kusanagi Y, Hamada K, Ochi H et al. (2001). Expression of E-cadherin and beta-catenin in primary and peritoneal metastatic ovarian carcinoma. Oncol Rep 8: 249–255.
Fujita T, Azuma Y, Fukuyama R, Hattori Y, Yoshida C, Koida M et al. (2004). Runx2 induces osteoblast and chondrocyte differentiation and enhances their migration by coupling with PI3K-Akt signaling. J Cell Biol 166: 85–95.
Harrington KS, Javed A, Drissi H, McNeil S, Lian JB, Stein JL et al. (2002). Transcription factors RUNX1/AML1 and RUNX2/Cbfa1 dynamically associate with stationary subnuclear domains. J Cell Sci 115: 4167–4176.
He DC, Nickerson JA, Penman S . (1990). Core filaments of the nuclear matrix. J Cell Biol 110: 569–580.
Inoue M, Ogawa H, Miyata M, Shiozaki H, Tanizawa O . (1992). Expression of E-cadherin in normal, benign, and malignant tissues of female genital organs. Am J Clin Pathol 98: 76–80.
Kirschnerova G, Tothova A, Babusikova O . (2006). Amplification of AML1 gene in association with karyotype, age and diagnosis in acute leukemia patients. Neoplasma 53: 150–154.
Levanon D, Bernstein Y, Negreanu V, Ghozi MC, Bar-Am I, Aloya R et al. (1996). A large variety of alternatively spliced and differentially expressed mRNAs are encoded by the human acute myeloid leukemia gene AML1. DNA Cell Biol 15: 175–185.
Liu YN, Lee WW, Wang CY, Chao TH, Chen Y, Chen JH . (2005). Regulatory mechanisms controlling human E-cadherin gene expression. Oncogene 24: 8277–8290.
Mikhail FM, Sinha KK, Saunthararajah Y, Nucifora G . (2006). Normal and transforming functions of RUNX1: a perspective. J Cell Physiol 207: 582–593.
Miyoshi H, Ohira M, Shimizu K, Mitani K, Hirai H, Imai T et al. (1995). Alternative splicing and genomic structure of the AML1 gene involved in acute myeloid leukemia. Nucleic Acids Res 23: 2762–2769.
Molinari E, Gilman M, Natesan S . (1999). Proteasome-mediated degradation of transcriptional activators correlates with activation domain potency in vivo. EMBO J 18: 6439–6447.
Orban TI, Olah E . (2003). Emerging roles of BRCA1 alternative splicing. Mol Pathol 56: 191–197.
Robinson HM, Broadfield ZJ, Cheung KL, Harewood L, Harris RL, Jalali GR et al. (2003). Amplification of AML1 in acute lymphoblastic leukemia is associated with a poor outcome. Leukemia 17: 2249–2250.
Sun L, Vitolo M, Passaniti A . (2001). Runt-related gene 2 in endothelial cells: inducible expression and specific regulation of cell migration and invasion. Cancer Res 61: 4994–5001.
Tanaka T, Kurokawa M, Ueki K, Tanaka K, Imai Y, Mitani K et al. (1996). The extracellular signal-regulated kinase pathway phosphorylates AML1, an acute myeloid leukemia gene product, and potentially regulates its transactivation ability. Mol Cell Biol 16: 3967–3979.
Whitley BR, Palmieri D, Twerdi CD, Church FC . (2004). Expression of active plasminogen activator inhibitor-1 reduces cell migration and invasion in breast and gynecological cancer cells. Exp Cell Res 296: 151–162.
Yamagata T, Maki K, Mitani K . (2005). Runx1/AML1 in normal and abnormal hematopoiesis. Int J Hematol 82: 1–8.
Yamaguchi Y, Kurokawa M, Imai Y, Izutsu K, Asai T, Ichikawa M et al. (2004). AML1 is functionally regulated through p300-mediated acetylation on specific lysine residues. J Biol Chem 279: 15630–15638.
Zaidi SK, Sullivan AJ, van Wijnen AJ, Stein JL, Stein GS, Lian JB . (2002). Integration of Runx and Smad regulatory signals at transcriptionally active subnuclear sites. Proc Natl Acad Sci USA 99: 8048–8053.
Zeng C, McNeil S, Pockwinse S, Nickerson J, Shopland L, Lawrence JB et al. (1998). Intranuclear targeting of AML/CBFalpha regulatory factors to nuclear matrix-associated transcriptional domains. Proc Natl Acad Sci USA 95: 1585–1589.
Zhang YW, Bae SC, Huang G, Fu YX, Lu J, Ahn MY et al. (1997). A novel transcript encoding an N-terminally truncated AML1/PEBP2 alphaB protein interferes with transactivation and blocks granulocytic differentiation of 32Dcl3 myeloid cells. Mol Cell Biol 17: 4133–4145.
Acknowledgements
This work was supported by the NCI P50 CA083639, P30 CA16672 and P01 CA64602 to GBM.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Nanjundan, M., Zhang, F., Schmandt, R. et al. Identification of a novel splice variant of AML1b in ovarian cancer patients conferring loss of wild-type tumor suppressive functions. Oncogene 26, 2574–2584 (2007). https://doi.org/10.1038/sj.onc.1210067
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.onc.1210067
Keywords
This article is cited by
-
mTOR in health and in sickness
Journal of Molecular Medicine (2015)