Abstract
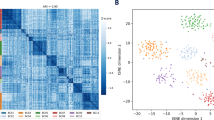
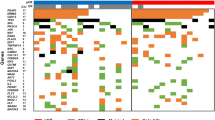
A series of studies have been published that evaluate the chromosomal copy number changes of different tumor classes using array comparative genomic hybridization (array CGH); however, the chromosomal aberrations that distinguish the different tumor classes have not been fully characterized. Therefore, we performed a meta-analysis of different array CGH data sets in an attempt to classify samples tested across different platforms. As opposed to RNA expression, a common reference is used in dual channel CGH arrays: normal human DNA, theoretically facilitating cross-platform analysis. To this aim, cell line and primary cancer data sets from three different dual channel array CGH platforms obtained by four different institutes were integrated. The cell line data were used to develop preprocessing methods, which performed noise reduction and transformed samples into a common format. The transformed array CGH profiles allowed perfect clustering by cell line, but importantly not by platform or institute. The same preprocessing procedures used for the cell line data were applied to data from 373 primary tumors profiled by array CGH, including controls. Results indicated that there is no apparent feature related to the institute or platform and that array CGH allows for unambiguous cross-platform meta-analysis. Major clusters with common tissue origin were identified. Interestingly, tumors of hematopoietic and mesenchymal origins cluster separately from tumors of epithelial origin. Therefore, it can be concluded that chromosomal aberrations of tumors from hematopoietic and mesenchymal origin versus tumors of epithelial origin are distinct, and these differences can be picked up by meta-analysis of array CGH data. This suggests the possibility of prospectively using combined analysis of diverse copy number data sets for cancer subtype classification.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
References
Albertson DG, Ylstra B, Segraves R, Collins C, Dairkee SH, Kowbel D et al. (2000). Quantitative mapping of amplicon structure by array CGH identifies CYP as a candidate oncogene. Nat Genet 25: 144–146.
Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C et al. (2001). Minimum information about a microarray experiment (MIAME) – toward standards for microarray data. Nat Genet 29: 365–371.
Douglas EJ, Fiegler H, Rowan A, Halford S, Bicknell DC, Bodmer W et al. (2004). Array comparative genomic hybridization analysis of colorectal cancer cell lines and primary carcinomas. Cancer Res 64: 4817–4825.
Edgar R, Domrachev M, Lash AE . (2002). Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30: 207–210.
Eisen MB, Spellman PT, Brown PO, Botstein D . (1998). Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95: 14863–14868.
Jong K, Marchiori E, Meijer G, Vaart AV, Ylstra B . (2004). Breakpoint identification and smoothing of array comparative genomic hybridization data. Bioinformatics 20: 3636–3637.
Jonsson G, Naylor TL, Vallon-Christersson J, Staaf J, Huang J, Ward MR et al. (2005). Distinct genomic profiles in hereditary breast tumors identified by array-based comparative genomic hybridization. Cancer Res 65: 7612–7621.
Linn SC, West RB, Pollack JR, Zhu S, Hernandez-Boussard T, Nielsen TO et al. (2003). Gene expression patterns and gene copy number changes in dermatofibrosarcoma protuberans. Am J Pathol 163: 2383–2395.
Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D et al. (2005). MicroRNA expression profiles classify human cancers. Nature 435: 834–838.
Michiels S, Koscielny S, Hill C . (2005). Prediction of cancer outcome with microarrays: a multiple random validation strategy. Lancet 365: 488–492.
Mitelman F, Johansson B, Mertens F . (2004). Fusion genes and rearranged genes as a linear function of chromosome aberrations in cancer. Nat Genet 36: 331–334.
Nakao K, Mehta KR, Fridlyand J, Moore DH, Jain AN, Lafuente A et al. (2004). High-resolution analysis of DNA copy number alterations in colorectal cancer by array-based comparative genomic hybridization. Carcinogenesis 25: 1345–1357.
Oostlander AE, Meijer GA, Ylstra B . (2004). Microarray-based comparative genomic hybridization and its applications in human genetics. Clin Genet 66: 488–495.
Pinkel D, Albertson DG . (2005). Array comparative genomic hybridization and its applications in cancer. Nat Genet 37 (Suppl): S11–S17.
Pollack JR, Perou CM, Alizadeh AA, Eisen MB, Pergamenschikov A, Williams CF et al. (1999). Genome-wide analysis of DNA copy-number changes using cDNA microarrays. Nat Genet 23: 41–46.
Pollack JR, Sorlie T, Perou CM, Rees CA, Jeffrey SS, Lonning PE et al. (2002). Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. Proc Natl Acad Sci USA 99: 12963–12968.
Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D et al. (2004). Large-scale meta-analysis of cancer microarray data identifies common transcriptional profiles of neoplastic transformation and progression. Proc Natl Acad Sci USA 101: 9309–9314.
Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N et al. (2003). TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34: 374–378.
Schreurs MW, Hermsen MA, Geltink RI, Scholten KB, Brink AA, Kueter EW et al. (2005). Genomic stability and functional activity may be lost in telomerase-transduced human CD8+ T lymphocytes. Blood 106: 2663–2670.
Snijders AM, Fridlyand J, Mans DA, Segraves R, Jain AN, Pinkel D et al. (2003a). Shaping of tumor and drug-resistant genomes by instability and selection. Oncogene 22: 4370–4379.
Snijders AM, Nowak N, Segraves R, Blackwood S, Brown N, Conroy J et al. (2001). Assembly of microarrays for genome-wide measurement of DNA copy number. Nat Genet 29: 263–264.
Snijders AM, Nowee ME, Fridlyand J, Piek JM, Dorsman JC, Jain AN et al. (2003b). Genome-wide-array-based comparative genomic hybridization reveals genetic homogeneity and frequent copy number increases encompassing CCNE1 in fallopian tube carcinoma. Oncogene 22: 4281–4286.
Snijders AM, Schmidt BL, Fridlyand J, Dekker N, Pinkel D, Jordan RC et al. (2005). Rare amplicons implicate frequent deregulation of cell fate specification pathways in oral squamous cell carcinoma. Oncogene 24: 4232–4242.
van Dekken H, Paris PL, Albertson DG, Alers JC, Andaya A, Kowbel D et al. (2004). Evaluation of genetic patterns in different tumor areas of intermediate-grade prostatic adenocarcinomas by high-resolution genomic array analysis. Genes Chromosomes Cancer 39: 249–256.
van den IJssel P, Tijssen M, Chin SF, Eijk P, Carvalho B, Hopmans E et al. (2005). Human and mouse oligonucleotide-based array CGH. Nucleic Acids Res 33: e192.
Weiss MM, Kuipers EJ, Postma C, Snijders AM, Siccama I, Pinkel D et al. (2003). Genomic profiling of gastric cancer predicts lymph node status and survival. Oncogene 22: 1872–1879.
Ylstra B, van den IJssel P, Carvalho B, Brakenhoff RH, Meijer GA . (2006). BAC to the future! or oligonucleotides: a perspective for micro array comparative genomic hybridization (array CGH). Nucleic Acids Res 34: 445–450.
Zardo G, Tiirikainen MI, Hong C, Misra A, Feuerstein BG, Volik S et al. (2002). Integrated genomic and epigenomic analyses pinpoint biallelic gene inactivation in tumors. Nat Genet 32: 453–458.
Acknowledgements
We thank Tony Cox, Ian Stalker (Sanger Institute, UK) and Nir Gamliel (Compugen USA, San Jose, CA, USA) for mapping the oligos into ENSEMBL. In addition, we would like to thank Cindy Postma and Erik Hopmans for their help with array hybridizations. We wish to thank Dr Stephen Ethier for the SUM cell lines and Dr Morag McCallum for the VP cell lines. SFC and CC are funded by Cancer Research UK. SFC was furthermore funded by COST-STSM B19. We furthermore acknowledge funding by the EU-sixth framework project: DISMAL, Contract No.: LSHC-CT-2005-018911. The work on the primary colon tumors at VUMC was supported by the Dutch Cancer Society Grant KWF-VU 02-2618.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc).
Rights and permissions
About this article
Cite this article
Jong, K., Marchiori, E., van der Vaart, A. et al. Cross-platform array comparative genomic hybridization meta-analysis separates hematopoietic and mesenchymal from epithelial tumors. Oncogene 26, 1499–1506 (2007). https://doi.org/10.1038/sj.onc.1209919
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.onc.1209919
Keywords
This article is cited by
-
Platform comparisons for identification of breast cancers with a BRCA-like copy number profile
Breast Cancer Research and Treatment (2013)
-
Wavelet-based identification of DNA focal genomic aberrations from single nucleotide polymorphism arrays
BMC Bioinformatics (2011)
-
Genome-wide comparison of paired fresh frozen and formalin-fixed paraffin-embedded gliomas by custom BAC and oligonucleotide array comparative genomic hybridization: facilitating analysis of archival gliomas
Acta Neuropathologica (2011)
-
An algorithm for classifying tumors based on genomic aberrations and selecting representative tumor models
BMC Medical Genomics (2010)
-
High-resolution DNA analysis of human embryonic stem cell lines reveals culture-induced copy number changes and loss of heterozygosity
Nature Biotechnology (2010)