Abstract
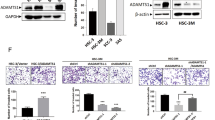
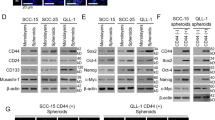
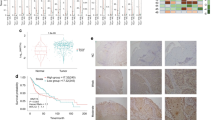
To identify genes that could potentially serve as molecular therapeutic markers for human head and neck cancer (HNC), we employed differential display analysis to compare the gene expression profiles between HNC and histopathologically normal epithelial tissues. Using reverse transcription–polymerase chain reaction and Western blot analysis, desmoglein 3 (DSG3) was identified as being differentially expressed at both the RNA and protein levels. Of 56 patients assayed, 34 (61%) had overexpression of DSG3, which correlated statistically with T stage (P=0.009), N stage (P=0.047), overall stage (P=0.011), tumor depth (P=0.009) and extracapsular spread in lymph nodes (P=0.044), suggesting that DSG3 participates in carcinogenesis of HNC. Consistent with the clinical findings, inhibition of DSG3 by RNA interference (RNAi) significantly reduced cell growth and colony formation to 57–21% in three HNC cell lines. Use of an in vitro wound healing and Matrigel invasion assays, we found that cell migration and invasive ability were also inhibited to 30–48% in three cell lines tested. An in vivo xenograft study showed that administration of DSG3-RNAi plasmid significantly inhibited tumor growth for 2 months in BALB/C nude mice. In conclusion, DSG3 is identified overexpressed in HNC, with the degree of overexpression associated with clinicopathologic features of the tumor. Inhibition of DSG3 significantly suppresses carcinogenic potential in cellular and in vivo animal studies. These findings suggest that DSG3 is a potential molecular target in the development of adjuvant therapy for HNC.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout









Similar content being viewed by others
References
Amagai M, Klaus-Kovtun V, Stanley JR . (1991). Autoantibodies against a novel epithelial cadherin in pemphigus vulgaris, a disease of cell adhesion. Cell 67: 869–877.
Amagai M, Nishikawa T, Nousari HC, Anhalt GJ, Hashimoto T . (1998). Antibodies against desmoglein 3 (pemphigus vulgaris antigen) are present in sera from patients with paraneoplastic pemphigus and cause acantholysis in vivo in neonatal mice. J Clin Invest 102: 775–782.
Chang JT, Chen IH, Liao CT, Wang HM, Hsu YM, Hung KF et al. (2002). A reverse transcription comparative real-time PCR method for quantitative detection of angiogenic growth factors in head and neck cancer patients. Clin Biochem 35: 591–596.
Chang JT, Lu YC, Chen YJ, Tseng CP, Chen YL, Fang CW et al. (2006). hTERT phosphorylation by PKC is essential for telomerase holoprotein integrity and enzyme activity in head neck cancer cells. Br J Cancer 94: 870–878.
Chang JT, Wang HM, Chang KW, Chen WH, Wen MC, Hsu YM et al. (2005). Identification of differentially expressed genes in oral squamous cell carcinoma (OSCC): overexpression of NPM, CDK1 and NDRG1 and underexpression of CHES1. Int J Cancer 114: 942–949.
Chung CH, Parker JS, Karaca G, Wu J, Funkhouser WK, Moore D et al. (2004). Molecular classification of head and neck squamous cell carcinomas using patterns of gene expression. Cancer Cell 5: 489–500.
Clayman GL, Lippman SM, Laramore GE, Hong WK . (1996). Cancer Medicine In: Holland JF, Frei E, Bast RC, Kufe DW, Morton DL, Weichselbaum R (eds). Williams and Wilkins: Philadelphia, pp. 1645–1709.
Decker J, Goldstein JC . (1982). Risk factors in head and neck cancer. N Engl J Med 306: 1151–1155.
Garrod D, Merritt AJ, Nie Z . (2002). Desmosomal cadherins. Curr Opin Cell Biol 14: 537–545.
Gordon LA, Mulligan KT, Maxwell-Jones H, Adams M, Walker RA, Jones JL . (2003). Breast cell invasive potential relates to the myoepithelial phenotype. Int J Cancer 106: 8–16.
Green KJ, Gaudry CA . (2000). Are desmosomes more than tethers for intermediate filaments? Nat Rev Mol Cell Biol 1: 208–216.
Hanakawa Y, Amagai M, Shirakata Y, Yahata Y, Tokumaru S, Yamasaki K et al. (2002a). Differential effects of desmoglein 1 and desmoglein 3 on desmosome formation. J Invest Dermatol 119: 1231–1236.
Hanakawa Y, Matsuyoshi N, Stanley JR . (2002b). Expression of desmoglein 1 compensates for genetic loss of desmoglein 3 in keratinocyte adhesion. J Invest Dermatol 119: 27–31.
Harada H, Iwatsuki K, Ohtsuka M, Han GW, Kaneko F . (1996). Abnormal desmoglein expression by squamous cell carcinoma cells. Acta Dermatol Venereol 76: 417–420.
Imai K, Kumagai S, Nakagawa K, Yamamoto E, Nakanishi I, Okada Y . (1995). Immunolocalization of desmoglein and intermediate filaments in human oral squamous cell carcinomas. Head Neck 17: 204–212.
Krunic AL, Garrod DR, Smith NP, Orchard GS, Cvijetic OB . (1996). Differential expression of desmosomal glycoproteins in keratoacanthoma and squamous cell carcinoma of the skin: an immunohistochemical aid to diagnosis. Acta Dermatol Venereol 76: 394–398.
Liao CT, Chang JT, Wang HM, Chen IH, Lin CY, Chen TM et al. (2004). Telomerase as an independent prognostic factor in head and neck squamous cell carcinoma. Head Neck 26: 504–512.
Liao SK, Perng YP, Shen YC, Chung PJ, Chang YS, Wang CH . (1998). Chromosomal abnormalities of a new nasopharyngeal carcinoma cell line (NPC-BM1) derived from a bone marrow metastatic lesion. Cancer Genet Cytogenet 103: 52–58.
Vokes EE, Weichselbaum RR, Lippman SM, Hong WK . (1993). Head and neck cancer. N Eng J Med 328: 184–194.
Yang CC, Tu SF, Chang RC, Hsu SF, Wu CH . (2001). In vitro cellular response of retinoic acid treated human oral cancer cell lines. Zhonghua Yi Xue Za Zhi (Taipei) 64: 357–363.
Yin T, Green KJ . (2004). Regulation of desmosome assembly and adhesion. Semin Cell Dev Biol 15: 665–677.
Acknowledgements
This work was supported by Chang Gung Memorial Hospital Grant (CMRPD32017 and CMRPD140141) and National Science Research Grant of Taiwan (NSC 94-2745-B-182-004-URD). We thank Mary Jeanne Buttrey, MD, for correction of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chen, YJ., Chang, J., Lee, L. et al. DSG3 is overexpressed in head neck cancer and is a potential molecular target for inhibition of oncogenesis. Oncogene 26, 467–476 (2007). https://doi.org/10.1038/sj.onc.1209802
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.onc.1209802
Keywords
This article is cited by
-
Drivers of plateau adaptability in cashmere goats revealed by genomic and transcriptomic analyses
BMC Genomics (2023)
-
Autoantigen microarrays reveal myelin basic protein autoantibodies in morphea
Journal of Translational Medicine (2022)
-
Oncoproteomic and gene expression analyses identify prognostic biomarkers for second primary malignancy in patients with head and neck squamous cell carcinoma
Modern Pathology (2019)
-
The desmosomal cadherin desmoglein-3 acts as a keratinocyte anti-stress protein via suppression of p53
Cell Death & Disease (2019)
-
The Endogenous GRP78 Interactome in Human Head and Neck Cancers: A Deterministic Role of Cell Surface GRP78 in Cancer Stemness
Scientific Reports (2018)