Abstract
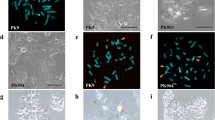
A gene critical to esophageal cancer has been identified. Functional studies using microcell-mediated chromosome transfer of intact and truncated donor chromosomes 3 into an esophageal cancer cell line and nude mouse tumorigenicity assays were used to identify a 1.61 Mb tumor suppressive critical region (CR) mapping to chromosome 3p14.2. This CR is bounded by D3S1600 and D3S1285 microsatellite markers. One candidate tumor suppressor gene, ADAMTS9, maps to this CR. Further studies showed normal expression levels of this gene in tumor-suppressed microcell hybrids, levels that were much higher than observed in the recipient cells. Complete loss or downregulation of ADAMTS9 gene expression was found in 15 out of 16 esophageal carcinoma cell lines. Promoter hypermethylation was detected in the cell lines that do not express this gene. Re-expression of ADAMTS9 was observed after demethylation drug treatment, confirming that hypermethylation is involved in gene downregulation. Downregulation of ADAMTS9 was also found in 43.5 and 47.6% of primary esophageal tumor tissues from Hong Kong and from the high-risk region of Henan, respectively. Thus, this study identifies and provides functional evidence for a CR associated with tumor suppression on 3p14.2 and provides the first evidence that ADAMTS9, mapping to this region, may contribute to esophageal cancer development.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Chen WY, Baylin SB . (2005). Cell Cycle 4: 10–12.
Cheng Y, Poulos NE, Lung ML, Hampton G, Ou B, Lerman MI et al. (1998). Proc Natl Acad Sci USA 95: 3042–3047.
Cheung AL, Si HX, Wang LD, An JY, Tsao SW . (2005). Cancer Lett (in press).
Clark ME, Kelner GS, Turbeville LA, Boyer A, Arden KC, Maki RA . (2000). Genomics 67: 343–350.
Deng W, Tsao SW, Guan XY, Lucas JN, Si HX, Leung CS et al. (2004). Oncogene 23: 9090–9101.
Goyette MC, Cho K, Fasching CL, Levy DB, Kinzler KW, Paraskeva C et al. (1992). Mol Cell Biol 12: 1387–1395.
Hesson L, Bieche I, Krex D, Criniere E, Hoang-Xuan K, Maher ER et al. (2004). Oncogene 23: 2408–2419.
Horiguchi K, Tomizawa Y, Tosaka M, Ishiuchi S, Kurihara H, Mori M et al. (2003). Oncogene 22: 7862–7865.
Hu CP, Hsieh HG, Chien KY, Wang PY, Wang CI, Chen CY et al. (1984). J Natl Cancer Inst 72: 577–583.
Hu YC, Lam KY, Law SY, Wan TS, Ma ES, Kwong YL et al. (2002). Cancer Genet Cytogenet 135: 120–127.
Ko JM, Wong CP, Tang CM, Lau KW, Lung ML . (2001). Cancer Lett 170: 131–138.
Ko JM, Yau WL, Chan PL, Lung HL, Yang L, Lo PH et al. (2005). Genes Chromosomes Cancer 43: 284–293.
Li JY . (1982). Natl Cancer Inst Monogr 62: 113–120.
Lo PH, Chung Leung AC, Xiong W, Law S, Duh FM, Lerman MI et al. (2006). Cancer Lett 234: 184–192.
Lung HL, Bangarusamy DK, Xie D, Cheung AKL, Cheng Y, Kumaran MK et al. (2005). Oncogene 24: 6525–6532.
Lung HL, Cheng Y, Kumaran MK, Liu ET, Murakami Y, Chan CY et al. (2004). Int J Cancer 112: 628–635.
Mok CH, Chew EC, Riches DJ, Lee JC, Huang DP, Hadgis C et al. (1987). Anticancer Res 7: 409–415.
Pan QQ . (1989). Proc Chin Acad Med Sci Peking Union Med Coll 4: 52–57.
Porter S, Scott SD, Sassoon EM, Williams MR, Jones JL, Girling AC et al. (2004). Clin Cancer Res 10: 2429–2440.
Qin YR, Wang LD, Kwong D, Gao SS, Guan XY, Zhuang ZH et al. (2005). Zhonghua Bing Li Xue Za Zhi 34: 80–83.
Rimessi P, Gualandi F, Morelli C, Trabanelli C, Wu Q, Possati L et al. (1994). Oncogene 9: 3467–3474.
Rooney DE . (1986). In Human Cytogenetics: A Practical Approach. Czeoulkowski BM (ed). IRL Press: Washington, DC.
Sambrook J, Fritsch EF, Maniatis T . (1989). Molecular Cloning. Cold Spring Harbor Laboratory Press: NY.
Saxon PJ, Srivatsan ES, Leipzig GV, Sameshima JH, Stanbridge EJ . (1985). Mol Cell Biol 5: 140–146.
Sekine I, Sato M, Sunaga N, Toyooka S, Peyton M, Parsons R et al. (2005). Oncogene 24: 2735–2738.
Shimada Y, Imamura M, Wagata T, Yamaguchi N, Tobe T . (1992). Cancer 69: 277–284.
Shiomi H, Sugihara H, Kamitani S, Tokugawa T, Tsubosa Y, Okada K et al. (2003). Cancer Genet Cytogenet 147: 50–61.
Somerville RP, Longpre JM, Jungers KA, Engle JM, Ross M, Evanko S et al. (2003). J Biol Chem 278: 9503–9513.
Takahashi K, Kanazama H, Chan CT . (1990). Jpn J Oral Maxillofac Surg 36: 307–316.
Tanaka H, Shimizu M, Horikawa I, Kugoh H, Yokota J, Barrett JC et al. (1998). Genes Chromosomes Cancer 23: 123–133.
Tang JC, Wan TS, Wong N, Pang E, Lam KY, Law SY et al. (2001). Cancer Genet Cytogenet 124: 36–41.
Tischoff I, Markwarth A, Witzigmann H, Uhlmann D, Hauss J, Mirmohammadsadegh A et al. (2005). Int J Cancer 115: 684–689.
Uzawa N, Yoshida MA, Hosoe S, Oshimura M, Amagasa T, Ikeuchi T . (1998). Cancer Genet Cytogenet 107: 125–131.
Wang L, Li W, Wang X, Zhang C, Zhang T, Mao X et al. (1996). Oncogene 12: 699–703.
Xiong W, Zeng ZY, Xia JH, Xia K, Shen SR, Li XL et al. (2004). Cancer Res 64: 1972–1974.
Yang L, Leung AC, Ko JM, Lo PH, Tang JC, Srivastava G et al. (2005). Oncogene 24: 697–705.
Yang Y, Kost-Alimova M, Ingvarsson S, Qianhui Q, Kiss H, Szeles A et al. (2001). Proc Natl Acad Sci USA 98: 1136–1141.
Acknowledgements
We acknowledge the financial support of the Research Grants Council of the Hong Kong Special Administration Region, China to MLL (HKUST6106/00M).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lo, P., Leung, A., Kwok, C. et al. Identification of a tumor suppressive critical region mapping to 3p14.2 in esophageal squamous cell carcinoma and studies of a candidate tumor suppressor gene, ADAMTS9. Oncogene 26, 148–157 (2007). https://doi.org/10.1038/sj.onc.1209767
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.onc.1209767
Keywords
This article is cited by
-
ADAMTS9-AS1 inhibits tumor growth and drug resistance in clear cell renal cell carcinoma via recruiting HuR to enhance ADAMTS9 mRNA stability
Cancer Nanotechnology (2023)
-
DNMT3A-mediated silence in ADAMTS9 expression is restored by RNF180 to inhibit viability and motility in gastric cancer cells
Cell Death & Disease (2021)
-
Aberrant DNA methylation of ADAMTS16 in colorectal and other epithelial cancers
BMC Cancer (2018)
-
The ADAMTS (A Disintegrin and Metalloproteinase with Thrombospondin motifs) family
Genome Biology (2015)
-
A new tumor suppressor LncRNA ADAMTS9-AS2 is regulated by DNMT1 and inhibits migration of glioma cells
Tumor Biology (2014)