Abstract
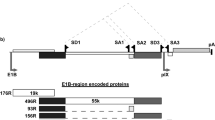
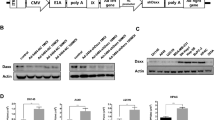
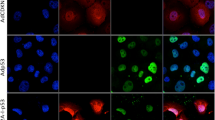
The tumor suppressor protein, p53, plays a critical role in viro-oncology. However, the role of p53 in adenoviral replication is still poorly understood. In this paper, we have explored further the effect of p53 on adenoviral replicative lysis. Using well-characterized cells expressing a functional p53 (A549, K1neo, RKO) and isogenic derivatives that do not (K1scx, RKOp53.13), we show that virus replication, late virus protein expression and both wtAd5 and ONYX-015 virus-induced cell death are impaired in cells deficient in functional p53. Conversely, by transfecting p53 into these and other cells (IIICF/c, HeLa), we increase late virus protein expression and virus yield. We also show, using reporter assays in IIICF/c, HeLa and K1scx cells, that p53 can cooperate with E1a to enhance transcription from the major late promoter of the virus. Late viral protein production is enhanced by exogenous p53. Taken together, our data suggest that functional p53 can promote the adenovirus (Ad) lytic cycle. These results have implications for the use of Ad mutants that are defective in p53 degradation, such as ONYX-015, as agents for the treatment of cancers.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Barker DD, Berk AJ . (1987). Virology 156: 107–121.
Bischoff JR, Kirn DH, Williams A, Heise C, Horn S, Muna M et al. (1996). Science 274: 373–376.
Braithwaite A, Nelson C, Skulimowski A, McGovern J, Pigott D, Jenkins J . (1990). Virology 177: 595–605.
Braithwaite AW, Murray JD, Bellett AJ . (1981). J Virol 39: 331–340.
Braithwaite AW, Sturzbecher HW, Addison C, Palmer C, Rudge K, Jenkins JR . (1987). Nature 329: 458–460.
Dix BR, Edwards SJ, Braithwaite AW . (2001). J Virol 75: 5443–5447.
Dix BR, O'Carroll SJ, Myers CJ, Edwards SJ, Braithwaite AW . (2000). Cancer Res 60: 2666–2672.
Edwards SJ, Dix BR, Myers CJ, Dobson-Le D, Huschtscha L, Hibma M et al. (2002). J Virol 76: 12483–12490.
Geoerger B, Grill J, Opolon P, Morizet J, Aubert G, Terrier-Lacombe MJ et al. (2002). Cancer Res 62: 764–772.
Goodrum FD, Ornelles DA . (1998). J Virol 72: 9479–9490.
Goodrum FD, Shenk T, Ornelles DA . (1996). J Virol 70: 6323–6335.
Hall AR, Dix BR, O'Carroll SJ, Braithwaite AW . (1998). Nat Med 4: 1068–1072.
Hann B, Balmain A . (2003). J Virol 77: 11588–11595.
Harada JN, Berk AJ . (1999). J Virol 73: 5333–5344.
Heise C, Sampson-Johannes A, Williams A, McCormick F, Von Hoff DD, Kirn DH . (1997). Nat Med 3: 639–645.
Hobom U, Dobbelstein M . (2004). J Virol 78: 7685–7697.
Holm PS, Bergmann S, Jurchott K, Lage H, Brand K, Ladhoff A et al. (2002). J Biol Chem 277: 10427–10434.
Jackson P, Ridgway P, Rayner J, Noble J, Braithwaite A . (1994). Biochem Biophys Res Commun 203: 133–140.
Jones N, Shenk T . (1978). Cell 13: 181–188.
Jones N, Shenk T . (1979). Proc Natl Acad Sci USA 76: 3665–3669.
Koch P, Gatfield J, Lober C, Hobom U, Lenz-Stoppler C, Roth J et al. (2001). Cancer Res 61: 5941–5947.
Kohno K, Izumi H, Uchiumi T, Ashizuka M, Kuwano M . (2003). BioEssays 25: 691–698.
Kruijer W, van Schaik FM, Sussenbach JS . (1981). Nucleic Acids Res 9: 4439–4457.
Lee H, Kim J, Lee B, Chang JW, Ahn J, Park JO et al. (2000). Int J Cancer 88: 454–463.
Lehman TA, Bennett WP, Metcalf RA, Welsh JA, Ecker J, Modali RV et al. (1991). Cancer Res 51: 4090–4096.
Morris GF, Mathews MB . (1991). J Virol 65: 6397–6406.
Nemunaitis J, Ganly I, Khuri F, Arseneau J, Kuhn J, McCarty T et al. (2000). Cancer Res 60: 6359–6366.
Nemunaitis J, Khuri F, Ganly I, Arseneau J, Posner M, Vokes E et al. (2001). J Clin Oncol 19: 289–298.
O'Carroll SJ, Hall AR, Myers CJ, Braithwaite AW, Dix BR . (2000). Biotechniques 28: 408–410.
O'Shea CC, Johnson L, Bagus B, Choi S, Nicholas C, Shen A et al. (2004). Cancer Cell 6: 611–623.
Ory K, Legros Y, Auguin C, Soussi T . (1994). EMBO J 13: 3496–3504.
Parks CL, Shenk T . (1997). J Virol 71: 9600–9607.
Pomerantz J, Schreiber-Agus N, Liegeois NJ, Silverman A, Alland L, Chin L et al. (1998). Cell 92: 713–723.
Prives C, Hall PA . (1999). J Pathol 187: 112–126.
Querido E, Blanchette P, Yan Q, Kamura T, Morrison M, Boivin D et al. (2001a). Genes Dev 15: 3104–3117.
Querido E, Morrison MR, Chu-Pham-Dang H, Thirlwell SW, Boivin D, Branton PE . (2001b). J Virol 75: 699–709.
Rauen KA, Sudilovsky D, Le JL, Chew KL, Hann B, Weinberg V et al. (2002). Cancer Res 62: 3812–3818.
Ries SJ, Brandts CH, Chung AS, Biederer CH, Hann BC, Lipner EM et al. (2000). Nat Med 6: 1128–1133.
Rogan EM, Bryan TM, Hukku B, Maclean K, Chang AC, Moy EL et al. (1995). Mol Cell Biol 15: 4745–4753.
Rogulski KR, Freytag SO, Zhang K, Gilbert JD, Paielli DL, Kim JH et al. (2000). Cancer Res 60: 1193–1196.
Rothmann T, Hengstermann A, Whitaker NJ, Scheffner M, Zur Hausen H . (1998). J Virol 72: 9470–9478.
Sarnow P, Sullivan CA, Levine AJ . (1982). Virology 120: 510–517.
Sauthoff H, Pipiya T, Heitner S, Chen S, Norman R, Rom W et al. (2002). Hum Gene Ther 13: 1859–1871.
Scheffner M, Munger K, Byrne J, Howley P . (1991). Proc Natl Acad Sci USA 88: 5523–5527.
Shenk T . (1996) In: Fields B, Knipe D, Howley P, Chanock R, Melnick J, Monath T, Roizman B, Strauss S (eds). Virology vol. 2. Lippincott-Raven: Philadelphia. pp 2111–2148.
Sleigh MJ . (1986). Anal Biochem 156: 251–256.
Slichenmyer WJ, Nelson WG, Slebos RJ, Kastan MB . (1993). Cancer Res 53: 4164–4168.
Stott FJ, Bates S, James MC, McConnell BB, Starborg M, Brookes S et al. (1998). EMBO J 17: 5001–5014.
Weyer U, Doerfler W . (1985). EMBO J 4: 3015–3019.
Williams J, Young C, Austin P . (1974). Cold Spring Harbor Sympos Quant Biol 39: 427–437.
Wyllie FS, Haughton MF, Rowson JM, Wynford-Thomas D . (1999). Br J Cancer 79: 1111–1120.
Yew PR, Berk AJ . (1992). Nature 357: 82–85.
Zhang YF, Homer C, Edwards SJ, Hananeia L, Lasham A, Royds J et al. (2003). Oncogene 22: 2782–2794.
Acknowledgements
We thank Mike Kastan (Johns Hopkins, Baltimore) for the RKO pair of cells, Roger Reddel (CMRI, Sydney) for the IIICF/c p53 null human fibroblast cell line, Walter Doerfler (Institut fur Genetik, Cologne) for the MLPCAT plasmid, and Peter van der Vliet (University Medical Center, Utrecht) for the DBP antibody. We wish to thank Nicky Real, Deidre Dobson-Le, Craig Homer and Rhodri Harfoot for technical assistance. This work was supported by grants from the Royal Society and the Health Research Council of New Zealand.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Royds, J., Hibma, M., Dix, B. et al. p53 promotes adenoviral replication and increases late viral gene expression. Oncogene 25, 1509–1520 (2006). https://doi.org/10.1038/sj.onc.1209185
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.onc.1209185
Keywords
This article is cited by
-
Enhanced oncolytic adenoviral production by downregulation of death-domain associated protein and overexpression of precursor terminal protein
Scientific Reports (2021)
-
The viral tropism of two distinct oncolytic viruses, reovirus and myxoma virus, is modulated by cellular tumor suppressor gene status
Oncogene (2010)
-
Combinatory cytotoxic effects produced by E1B-55kDa-deleted adenoviruses and chemotherapeutic agents are dependent on the agents in esophageal carcinoma
Cancer Gene Therapy (2010)