Abstract
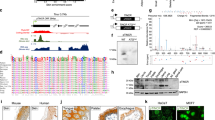
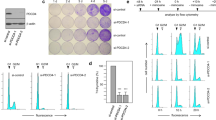
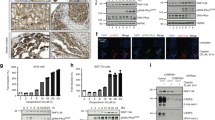
tRNA-isopentenyltransferase (tRNA-IPT) catalyses the addition of N6-isopentenyladenosine (i6A) on residue 37 of tRNA molecules that bind codons starting with uridine. Post-transcriptional modifications of tRNA molecules have been demonstrated to be essential in maintaining the correct reading frame of the translational machinery, thus improving fidelity and efficiency of protein synthesis. We show here that the human tRNA-isopentenyltransferase (TRIT1) gene encodes a complex pattern of mRNA variants through alternative splicing in both normal and tumor lung tissue and that the nonsense suppressor activity of tRNA-IPT is maintained only in the full-length mRNA isoform, as revealed by gene complementation in yeast. Expression of the full-length transcript was down-regulated 6–14-fold in lung adenocarcinomas as compared to normal lung tissue. A549 lung cancer cells transfected to express the functional TRIT1 gene formed significantly smaller colonies with reduced scattering on the edges and had only limited ability to induce tumors in nude mice. Our findings raise the possibility of TRIT1 as a candidate lung tumor suppressor.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
References
Berry MJ, Tujebajeva RM, Copeland PR, Xu XM, Carlson BA, Martin GW, III, Low SC, Mansell JB, Grundner-Culemann E, Harney JW, Driscoll DM and Hatfield DL . (2001). Biofactors, 14, 17–24.
Bjork GR, Durand JM, Hagervall TG, Leipuviene R, Lundgren HK, Nilsson K, Chen P, Qian Q and Urbonavicius J . (1999). FEBS Lett., 452, 47–51.
Bjornsti MA and Houghton PJ . (2004). Cancer Cell, 5, 519–523.
Caillet J and Droogmans L . (1988). J. Bacteriol., 170, 4147–4152.
Connolly DM and Winkler ME . (1989). J. Bacteriol., 171, 3233–3246.
Diaz I, Pedersen S and Kurland CG . (1987). Mol. Gen. Genet., 208, 373–376.
Dihanich ME, Najarian D, Clark R, Gillman EC, Martin NC and Hopper AK . (1987). Mol. Cell. Biol., 7, 177–184.
Dirheimer G, Baranowski W and Keith G . (1995). Biochimie, 77, 99–103.
Gehrke CW, Kuo KC, McCune RA, Gerhardt KO and Agris PF . (1982). J. Chromatogr., 230, 297–308.
Gietz RD and Schiestl RH . (1997). Methods Mol. Cell. Biol., 5, 255–269.
Gillman EC, Slusher LB, Martin NC and Hopper AK . (1991). Mol. Cell. Biol., 11, 2382–2390.
Golovko A, Hjalm G, Sitbon F and Nicander B . (2000). Gene, 258, 85–93.
Lemieux J, Lakowski B, Webb A, Meng Y, Ubach A, Bussiere F, Barnes T and Hekimi S . (2001). Genetics, 159, 147–157.
Leung HC, Chen Y and Winkler ME . (1997). J. Biol. Chem., 272, 13073–13083.
Low SC and Berry MJ . (1996). Trends Biochem. Sci., 21, 203–208.
Miller CO, Skoog F, von Saltza MH and Strong F . (1955). J. Am. Chem. Soc., 77, 1392–1393.
Mok MC . (1994) Mok DWS and Mok MC (eds). Cytokinins. Chemistry Activity, and Function. CRC Press: Boca Raton, pp 155–166.
Moustafa ME, Carlson BA, El Saadani MA, Kryukov GV, Sun QA, Harney JW, Hill KE, Combs GF, Feigenbaum L, Mansur DB, Burk RF, Berry MJ, Diamond AM, Lee BJ, Gladyshev VN and Hatfield DL . (2001). Mol. Cell. Biol., 21, 3840–3852.
Persson BC . (1993). Mol. Microbiol., 8, 1011–1016.
Persson BC, Esberg B, Olafsson O and Bjork GR . (1994). Biochimie, 76, 1152–1160.
Rozenski J, Crain PF and McCloskey JA . (1999). Nucleic Acids Res., 27, 196–197.
Sakakibara H and Takei K . (2002). J. Plant Growth Regul., 21, 17–23.
Takei K, Sakakibara H and Sugiyama T . (2001). J. Biol. Chem., 276, 26405–26410.
Urbonavicius J, Qian Q, Durand JM, Hagervall TG and Bjork GR . (2001). EMBO J., 20, 4863–4873.
Warner GJ, Berry MJ, Moustafa ME, Carlson BA, Hatfield DL and Faust JR . (2000). J. Biol. Chem., 275, 28110–28119.
Acknowledgements
We thank A Hopper for the gift of MT-8 and H57 yeast strains. This work was funded in part by grants from Associazione and Fondazione Italiana Ricerca Cancro (AIRC and FIRC) and FIRB, Italy.
Author information
Authors and Affiliations
Corresponding author
Additional information
Data deposition footnote: Sequence data are available from GenBank under accession numbers AY702933 to AY702947.
Rights and permissions
About this article
Cite this article
Spinola, M., Galvan, A., Pignatiello, C. et al. Identification and functional characterization of the candidate tumor suppressor gene TRIT1 in human lung cancer. Oncogene 24, 5502–5509 (2005). https://doi.org/10.1038/sj.onc.1208687
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.onc.1208687
Keywords
This article is cited by
-
Genetically regulated expression in late-onset Alzheimer’s disease implicates risk genes within known and novel loci
Translational Psychiatry (2021)
-
Disruption of the RNA modifications that target the ribosome translation machinery in human cancer
Molecular Cancer (2020)
-
Anti-cancer activities of cytokinin ribosides
Phytochemistry Reviews (2019)
-
N6-isopentenyladenosine dual targeting of AMPK and Rab7 prenylation inhibits melanoma growth through the impairment of autophagic flux
Cell Death & Differentiation (2018)
-
The mitochondrial epitranscriptome: the roles of RNA modifications in mitochondrial translation and human disease
Cellular and Molecular Life Sciences (2018)