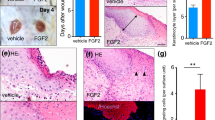
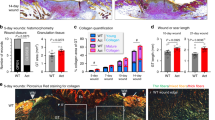
Abstract
The fibroblast growth factor-binding protein (FGF-BP) binds and activates FGF-1 and FGF-2, thereby contributing to tumor angiogenesis. In this study, we identified novel binding partners of FGF-BP, and we provide evidence for a role of this protein in epithelial repair processes. We show that expression of FGF-BP increases after injury to murine and human skin, in particular in keratinocytes. This upregulation is most likely achieved by major keratinocyte mitogens present at the wound site. Most importantly, we demonstrate that FGF-BP interacts with FGF-7, FGF-10, and with the recently identified FGF-22, and enhances the activity of low concentrations of ligand. Due to the important functions of FGF-7 and FGF-10 for repair of injured epithelia, our findings suggest that upregulation of FGF-BP expression after injury stimulates FGF activity at the wound site, thus enhancing the process of epithelial repair.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
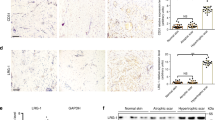
References
Aigner A, Butscheid M, Kunkel P, Krause E, Lamszus K, Wellstein A and Czubayko F . (2001). Int. J. Cancer, 92, 510–517.
Aigner A, Malerczyk C, Houghtling R and Wellstein A . (2000). Growth Factors, 18, 51–62.
Aigner A, Ray PE, Czubayko F and Wellstein A . (2002b). Histochem. Cell Biol., 117, 1–11.
Aigner A, Renneberg H, Bojunga J, Apel J, Nelson PS and Czubayko F . (2002a). Oncogene, 21, 5733–5742.
Beer HD, Florence C, Dammeier J, McGuire L, Werner S and Duan DR . (1997). Oncogene, 15, 2211–2218.
Beyer TA, Werner S, Dickson C and Grose R . (2003). Exp. Cell Res., 287, 228–236.
Boukamp P, Petrussevska RT, Breitkreutz D, Hornung J, Markham A and Fusenig NE . (1988). J. Cell Biol., 106, 761–771.
Brauchle M, Angermeyer K, Hubner G and Werner S . (1994). Oncogene, 9, 3199–3204.
Chen Y, Chou K, Fuchs E, Havran WL and Boismenu R . (2002). Proc. Natl. Acad. Sci. USA, 99, 14338–14343.
Chomczynski P and Sacchi N . (1987). Anal. Biochem., 162, 156–159.
Czubayko F, Liaudet-Coopman ED, Aigner A, Tuveson AT, Berchem GJ and Wellstein A . (1997). Nat. Med., 3, 1137–1140.
Czubayko F, Smith RV, Chung HC and Wellstein A . (1994). J. Biol. Chem., 269, 28243–28248.
Dell KR and Williams LT . (1992). J. Biol. Chem., 267, 21225–21229.
Finch PW and Rubin JS . (2004). Adv. Cancer Res., 91, 69–136.
Harris VK, Coticchia CM, Kagan BL, Ahmad S, Wellstein A and Riegel AT . (2000). J. Biol. Chem., 275, 10802–10811.
Igarashi M, Finch PW and Aaronson SA . (1998). J. Biol. Chem., 273, 13230–13235.
Johnson DE and Williams LT . (1993). Adv. Cancer Res., 60, 1–41.
Kurtz A, Aigner A, Cabal-Manzano RH, Butler RE, Hood DR, Sessions RB, Czubayko F and Wellstein A . (2004). Neoplasia, 6, 595–602.
Kurtz A, Wang HL, Darwiche N, Harris V and Wellstein A . (1997). Oncogene, 14, 2671–2681.
Liu X-H, Aigner A, Wellstein A and Ray PE . (2001). Kidney Int., 59, 1717–1728.
Marchese C, Chedid M, Dirsch OR, Csaky KG, Santanelli F, Latini C, LaRochelle WJ, Torrisi MR and Aaronson SA . (1995). J. Exp. Med., 182, 1369–1376.
Miki T, Bottaro DP, Fleming TP, Smith CL, Burgess WH, Chan AM and Aaronson SA . (1992). Proc. Natl. Acad. Sci. USA, 89, 246–250.
Mongiat M, Otto J, Oldershaw R, Ferrer F, Sato D and Iozzo RV . (2001). J. Biol. Chem., 276, 10263–10271.
Nakatake Y, Hoshikawa M, Asaki T, Kassai Y and Itoh N . (2001). Biochim. Biophys. Acta, 1517, 460–463.
Okamoto T, Tanaka Y, Kan M, Sakamoto A, Takada K and Sato JD . (1996). In vitro Cell. Dev. Biol. Anim., 32, 69–71.
Ornitz DM . (2000). BioEssays, 22, 108–112.
Ornitz DM and Itoh N . (2001). Genome Biol., 2, Reviews 2005.
Ornitz DM, Xu J, Colvin JS, McEwen DG, MacArthur CA, Coulier F, Gao G and Goldfarb M . (1996). J. Biol. Chem., 271, 15292–15297.
Orr-Urtreger A, Bedford MT, Burakova T, Arman E, Zimmer Y, Yayon A, Givol D and Lonai P . (1993). Dev. Biol., 158, 475–486.
Powers CJ, McLeskey SW and Wellstein A . (2000). Endocr. Relat. Cancer, 7, 165–197.
Rubin JS, Osada DP, Finch PW, Taylor WG, Rudikoff S and Aaronson SA . (1989). Proc. Natl. Acad. Sci. USA, 86, 802–806.
Sauter ER, Nesbit M, Tichansky D, Liu ZJ, Shirakawa T, Palazzo J and Herlyn M . (2001). Int. J. Cancer, 92, 374–381.
Shipley GD, Pittelkow MR, Wille Jr JJ, Scott RE and Moses HL . (1986). Cancer Res., 46, 2068–2071.
Steiling H and Werner S . (2003). Curr. Opin. Biotechnol., 14, 533–537.
Steiling H, Wustefeld T, Bugnon P, Brauchle M, Fassler R, Teupser D, Thiery J, Gordon JI, Trautwein C and Werner S . (2003). Oncogene, 22, 4380–4388.
Tagashira S, Harada H, Katsumata T, Itoh N and Nakatsuka M . (2001). Gene, 197, 399–404.
Tassi E, Al-Attar A, Aigner A, Swift MR, McDonnell K, Karavanov A and Wellstein A . (2001). J. Biol. Chem., 276, 40247–40253.
Werner S, Breeden M, Hübner G, Greenhalgh DG and Longaker MT . (1994b). J. Invest. Dermatol., 103, 469–473.
Werner S, Duan DS, de Vries C, Peters KG, Johnson DE and Williams LT . (1992). Mol. Cell. Biol., 12, 82–88.
Werner S and Grose R . (2003). Phys. Rev., 83, 835–870.
Werner S, Smola H, Liao X, Longaker MT, Krieg T, Hofschneider PH and Williams LT . (1994a). Science, 266, 819–822.
Werner S, Weinberg W, Liao X, Peters KG, Blessing M, Yuspa SH, Weiner RL and Williams LT . (1993). EMBO J., 12, 2635–2643.
Wu DQ, Kan MK, Sato GH, Okamoto T and Sato JD . (1991). J. Biol. Chem., 266, 16778–16785.
Acknowledgements
This work was supported by the Swiss National Science Foundation (grant No. 31-61358.00 to SW), the German Ministry for Education and Research (BMBF to SW and AG), and the ETH Zürich (to SW). We thank Christiane Born-Berclaz for excellent technical assistance, Dr Richard Grose and Tobias Beyer for the nontagged and HA-tagged FGF-22 cDNAs, Dr Clive Dickson for helpful suggestions, and Dr Jeffrey Rubin, NIH, Bethesda, for critically reading the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Beer, HD., Bittner, M., Niklaus, G. et al. The fibroblast growth factor binding protein is a novel interaction partner of FGF-7, FGF-10 and FGF-22 and regulates FGF activity: implications for epithelial repair. Oncogene 24, 5269–5277 (2005). https://doi.org/10.1038/sj.onc.1208560
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.onc.1208560
Keywords
This article is cited by
-
Fibroblast growth factor signaling in axons: from development to disease
Cell Communication and Signaling (2023)
-
Eldecalcitol (ED-71)-induced exosomal miR-6887-5p suppresses squamous cell carcinoma cell growth by targeting heparin-binding protein 17/fibroblast growth factor–binding protein-1 (HBp17/FGFBP-1)
In Vitro Cellular & Developmental Biology - Animal (2020)
-
Advances in surgical applications of growth factors for wound healing
Burns & Trauma (2019)
-
Fibroblast Growth Factor Binding Protein 3 (FGFBP3) impacts carbohydrate and lipid metabolism
Scientific Reports (2018)
-
RNA-Seq analysis of Gtf2ird1 knockout epidermal tissue provides potential insights into molecular mechanisms underpinning Williams-Beuren syndrome
BMC Genomics (2016)