Abstract
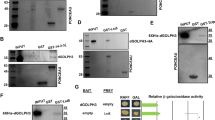
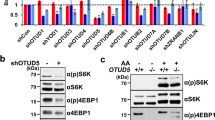
TRC8 encodes an E3-ubiquitin ligase disrupted in a family with hereditary renal cell carcinoma (RCC). We previously reported that Drosophila Trc8 (DTrc8) overexpression inhibits growth and that human and fly proteins interact with with the COP9 signalosome (CSN) subunit JAB1/CSN5. However, further mechanistic evidence linking DTrc8 growth suppression to CSN5 was lacking. Here, we show that haploinsufficiency of CSN5, or a T100I point mutation (CSN53), relieved growth suppression by DTrc8, whereas CSN51 (E160V) and CSN52 (G147D) mutations had no effect. The strength of yeast two-hybrid interactions between DTrc8 and CSN5 were in complete agreement with the observed phenotypes. DTrc8 overexpression resulted in elevated levels of CSN5 and CSN7, but had no effect on NEDD8-modified Cul-1. In contrast to CSN5, heterozygosity for CSN4null had no effect on the DTrc8 phenotype. We also looked for genetic interactions between DTrc8 and other MPN domain proteins in the CSN and 26S proteasome lid. CSN6 haploinsufficiency restored growth, whereas reduction of proteasome subunits RPN8 or RPN11 had no effect. DTrc8 expression increased the level of digitonin-extractable CSN complex, consistent with elevated levels of CSN5 and 7. Our genetic results confirm that DTrc8-induced growth suppression is CSN5 (and CSN6) dependent. While there was no obvious influence on CSN deneddylation activity, the increase in CSN subunits and holocomplex suggests that TRC8 modulates signalosome levels or compartmentalization.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Bianchi E, Denti S, Granata A, Bossi G, Geginat J, Villa A, Rogge L and Pardi R . (2000). Nature, 404, 617–621.
Chamovitz DA and Glickman M . (2002). Curr. Biol., 12, R232.
Chamovitz DA and Segal D . (2001). EMBO Rep., 2, 96–101.
Cohen AJ, Li FP, Berg S, Marchetto DJ, Tsai S, Jacobs SC and Brown RS . (1979). N. Engl. J. Med., 301, 592–595.
Cope GA and Deshaies RJ . (2003). Cell, 114, 663–671.
Cope GA, Suh GS, Aravind L, Schwarz SE, Zipursky SL, Koonin EV and Deshaies RJ . (2002). Science, 298, 608–611.
Freilich S, Oron E, Kapp Y, Nevo-Caspi Y, Orgad S, Segal D and Chamovitz DA . (1999). Curr. Biol., 9, 1187–1190.
Gemmill RM, Bemis LT, Lee JP, Sozen MA, Baron A, Zeng C, Erickson PF, Hooper JE and Drabkin HA . (2002). Oncogene, 21, 3507–3516.
Gemmill RM, West JD, Boldog F, Tanaka N, Robinson LJ, Smith DI, Li F and Drabkin HA . (1998). Proc. Natl. Acad. Sci. USA, 95, 9572–9577.
Glickman MH, Rubin DM, Coux O, Wefes I, Pfeifer G, Cjeka Z, Baumeister W, Fried VA and Finley D . (1998). Cell, 94, 615–623.
Kim M, Katayosc Y, Li Q, Rakkar AN, Li Z, Hwang SG, Katayose D, Trepel J, Cowan KH and Seth P . (1998). Biochem. Biophys. Res. Commun., 253, 672–677.
Kleemann R, Hausser A, Geiger G, Mischke R, Burger-Kentischer A, Flieger O, Johannes FJ, Roger T, Calandra T, Kapurniotu A, Grell M, Finkelmeier D, Brunner H and Bernhagen J . (2000). Nature, 408, 211–216.
Li FP, Decker HJ, Zbar B, Stanton Jr VP, Kovacs G, Seizinger BR, Aburatani H, Sandberg AA, Berg S and Hosoe S . (1993). Ann. Intern. Med., 118, 106–111.
Lorick KL, Jensen JP, Fang S, Ong AM, Hatakeyama S and Weissman AM . (1999). Proc. Natl. Acad. Sci. USA, 96, 11364–11369.
Lyapina S, Cope G, Shevchenko A, Serino G, Tsuge T, Zhou C, Wolf DA, Wei N, Shevchenko A and Deshaies RJ . (2001). Science, 292, 1382–1385.
Maytal-Kivity V, Reis N, Hofmann K and Glickman MH . (2002). BMC Biochem., 3, 28.
Mundt KE, Liu C and Carr AM . (2002). Mol. Biol. Cell, 13, 493–502.
Oron E, Mannervik M, Rencus S, Harari-Steinberg O, Neuman-Silberberg S, Segal D and Chamovitz DA . (2002). Development, 129, 4399–4409.
Schwechheimer C, Serino G, Callis J, Crosby WL, Lyapina S, Deshaies RJ, Gray WM, Estelle M and Deng XW . (2001). Science, 292, 1379–1382.
Suh GS, Poeck B, Chouard T, Oron E, Segal D, Chamovitz DA and Zipursky SL . (2002). Neuron, 33, 35–46.
Tomoda K, Kato JY, Tatsumi E, Takahashi T, Matsuo Y and Yoneda-Kato N . (2005). Blood, 105, 775–783.
Tomoda K, Kubota Y, Arata Y, Mori S, Maeda M, Tanaka T, Yoshida M, Yoneda-Kato N and Kato JY . (2002). J. Biol. Chem., 277, 2302–2310.
Wei N and Deng XW . (2003). Annu. Rev. Cell Dev. Biol., 19, 261–286.
Acknowledgements
We thank Drs Steven Beckendorf and CT Chien for providing multiple fly strains, including the CSN52 missense mutation. Statistical analysis was performed by Drs Anna Baron and Chan Zeng of the University of Colorado Cancer Center Biostatistics Core. We also thank D Kent and C Korch for helpful discussions during the course of this work. These studies were supported by an NIH grant (CA76035) to HAD and RMG, and from the Israel Science Foundation to DAC and DS.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Gemmill, R., Lee, J., Chamovitz, D. et al. Growth suppression induced by the TRC8 hereditary kidney cancer gene is dependent upon JAB1/CSN5. Oncogene 24, 3503–3511 (2005). https://doi.org/10.1038/sj.onc.1208509
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.onc.1208509